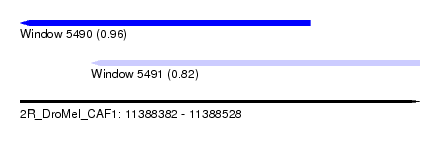
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,388,382 – 11,388,528 |
| Length | 146 |
| Max. P | 0.964396 |

| Location | 11,388,382 – 11,388,488 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -43.48 |
| Consensus MFE | -32.31 |
| Energy contribution | -32.73 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
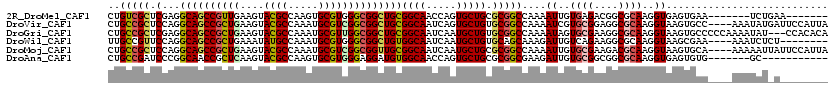
>2R_DroMel_CAF1 11388382 106 - 20766785 CUGUCGCUCGAGGCAGCCGUUGAAGUACGCCAAGUGCGUGGGCGGCUGCGGCAACCAGUGCUGCGCGGCCAAAAUUGUGAGACGGCGCAAGGUGAGUGAA-------UCUGAA------- ...(((((((..((.((((((....(((((.....)))))(((.((.((((((.....)))))))).)))..........))))))))....))))))).-------......------- ( -45.30) >DroVir_CAF1 6319 116 - 1 CUGCCGCUCCAGGCAGCCGCUGAAGUACGCCAAAUGCGUCGGCGGCUGCGGCAAUCAGUGCUGUGCGGCCAAAAUCGUGCGGAGGCGCAAGGUAAGUGCC----AAAUAUGAUUCCAUUA ..(((((.....(((((((((((....(((.....))))))))))))))((((.....))))..)))))...(((((((....(((((.......)))))----...)))))))...... ( -48.20) >DroGri_CAF1 5462 117 - 1 CUGCCGCUCGAGGCAGCCGCUGAAGUACGCCAAAUGCGUUGGCGGCUGCGGCAAUCAAUGCUGUGCGGCCAAAAUAGUGCGAAGGCGCAAGGUAAGUGCCCCCAAAAUAU---CCACACA ..(((((.(...(((((((((.....((((.....)))).)))))))))((((.....))))).))))).......((((....))))..(((....)))..........---....... ( -45.30) >DroWil_CAF1 5267 108 - 1 UUGCCGUUCCAGGCAGCCGCUGAAAUAUGCCAAAUGCGUGGGCGGCUGUGGCAAUCAAUGCUGUGCAGCAAAGAUUGUCAGAAGGCGCAAGGUAAGCGAA----AAAUCUCU-------- ..(((.(((...(((((((((....(((((.....))))))))))))))(((((((..((((....))))..))))))).))))))((.......))...----........-------- ( -40.50) >DroMoj_CAF1 5955 116 - 1 CUGCCGCUCCAGGCAGCCGCUGAAGUACGCCAAAUGCGUCGGCGGUUGCGGCAAUCAAUGCUGCGCGGCCAAAAUUGUGCGAAGACGCAAGGUAAGUGCA----AAAAAUUAUUCCAUUA ..(((((.....(((((((((((....(((.....))))))))))))))((((.....))))..))))).....(((..(.....(....)....)..))----)............... ( -42.60) >DroAna_CAF1 4608 102 - 1 CUGCCGAUCCCGGCAACCGCUCAAGUACGCCAAGUGCGUGGGAGGAUGUGGCAACCAGUGCUGCGCGGCGAAGAUUGUGCGGCGGCGCAAGGUGAGUGUG-------GC----------- .(((((....))))).((((((......((((.((.(......).)).)))).(((.((((((((((.(((...)))))).)))))))..)))))))).)-------..----------- ( -39.00) >consensus CUGCCGCUCCAGGCAGCCGCUGAAGUACGCCAAAUGCGUGGGCGGCUGCGGCAAUCAAUGCUGCGCGGCCAAAAUUGUGCGAAGGCGCAAGGUAAGUGCA____AAAUAU_A________ ..(((((.(...((((((((((....((((.....))))))))))))))((((.....))))).)))))....((..((((....))))..))........................... (-32.31 = -32.73 + 0.42)

| Location | 11,388,408 – 11,388,528 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.72 |
| Mean single sequence MFE | -49.48 |
| Consensus MFE | -42.29 |
| Energy contribution | -42.63 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
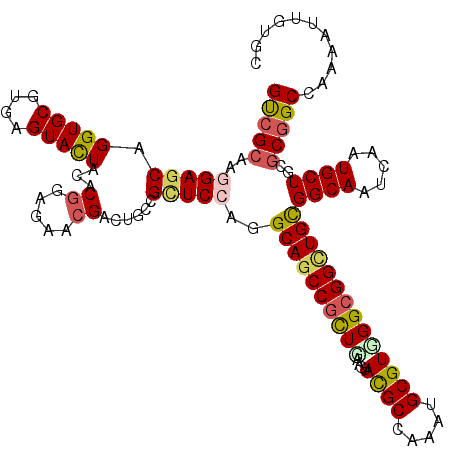
>2R_DroMel_CAF1 11388408 120 - 20766785 GUCGCAAGGAGCAGGUGCGCGAGUACUACACGGAGAACGACUGUCGCUCGAGGCAGCCGUUGAAGUACGCCAAGUGCGUGGGCGGCUGCGGCAACCAGUGCUGCGCGGCCAAAAUUGUGA .((((((.((((.(((((....)))))...((.....))......))))..((((((((((.(.((((.....)))).).)))))))((((((.....))))))...)))....)))))) ( -51.00) >DroVir_CAF1 6355 120 - 1 GUCGCAAGGAGCAGGUGCGUGAGUAUUACACGGAGAACGACUGCCGCUCCAGGCAGCCGCUGAAGUACGCCAAAUGCGUCGGCGGCUGCGGCAAUCAGUGCUGUGCGGCCAAAAUCGUGC ((((((.(((((.(((((....)))))....((((.....)).)))))))..(((((((((((....(((.....))))))))))))))((((.....)))).))))))........... ( -53.10) >DroGri_CAF1 5499 120 - 1 GCCGCAAGGAGCAGGUGCGAGAGUAUUACACGGAGAACGACUGCCGCUCGAGGCAGCCGCUGAAGUACGCCAAAUGCGUUGGCGGCUGCGGCAAUCAAUGCUGUGCGGCCAAAAUAGUGC ((((((...(((((((((....)))))...........((.((((.(....)(((((((((.....((((.....)))).))))))))))))).))..)))).))))))........... ( -49.40) >DroWil_CAF1 5295 120 - 1 GUCGGAAGGAGCAGGUGCGUGAGUAUUACACAGAGAACGAUUGCCGUUCCAGGCAGCCGCUGAAAUAUGCCAAAUGCGUGGGCGGCUGUGGCAAUCAAUGCUGUGCAGCAAAGAUUGUCA ...((((.(.((((((((....))))).(.....)......)))).))))..(((((((((....(((((.....))))))))))))))(((((((..((((....))))..))))))). ( -46.60) >DroMoj_CAF1 5991 120 - 1 GUCGCAAGGAGCAGGUGCGUGAGUACUACACGGAGAACGACUGCCGCUCCAGGCAGCCGCUGAAGUACGCCAAAUGCGUCGGCGGUUGCGGCAAUCAAUGCUGCGCGGCCAAAAUUGUGC ..((((((((((.(((((....)))))....((((.....)).))))))).((((((((((((....(((.....))))))))))))((((((.....))))))...)))....))))). ( -53.30) >DroAna_CAF1 4630 120 - 1 GUCGCAAGGAGCAGGUGCGCGAGUACUACACGGAGAACGACUGCCGAUCCCGGCAACCGCUCAAGUACGCCAAGUGCGUGGGAGGAUGUGGCAACCAGUGCUGCGCGGCGAAGAUUGUGC (((((..(.(((((((((....))))).((((.........(((((....))))).((.((((.((((.....)))).)))).)).))))........)))).))))))........... ( -43.50) >consensus GUCGCAAGGAGCAGGUGCGUGAGUACUACACGGAGAACGACUGCCGCUCCAGGCAGCCGCUGAAGUACGCCAAAUGCGUGGGCGGCUGCGGCAAUCAAUGCUGCGCGGCCAAAAUUGUGC (((((..(((((.(((((....)))))...((.....))......)))))..((((((((((....((((.....))))))))))))))((((.....))))..)))))........... (-42.29 = -42.63 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:37 2006