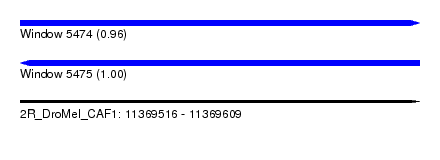
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,369,516 – 11,369,609 |
| Length | 93 |
| Max. P | 0.996056 |

| Location | 11,369,516 – 11,369,609 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 71.14 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -9.87 |
| Energy contribution | -11.31 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.958972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
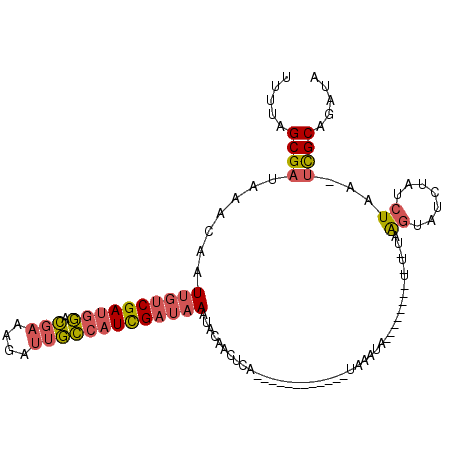
>2R_DroMel_CAF1 11369516 93 + 20766785 UAUCUGCGA-AUAGAUAGAUACUUAUACA------UAUAUUUA------------UGAGUUUUACUUAUCGAUGGCAAUCUUUCGUCCAUCGACAAUUGUUUAUCGCUAAAA .....((((-.(((((((..(((((((..------......))------------)))))........((((((((........).)))))))...)))))))))))..... ( -21.80) >DroSec_CAF1 2271 87 + 1 UAUCGGCGA-UUAGAUAGAUACUUA------------UAUUUA------------UGAGUUGUAUUUAUCGAUGGCAAUCUUUCGUCCAUCGACAAUUGUUUAUCGCUAAAA ....(((((-(((((((.(.(((((------------(....)------------)))))).))))))((((((((........).))))))).........)))))).... ( -22.60) >DroSim_CAF1 2265 87 + 1 UAUCUGCGA-UUAGAUAGAUACUUA------------UAUUUA------------UGAGUUGUAUUUAUCGAUGGCAAUCUUUCGUCCAUCGACAAUUGUUUAUCGCUAAAA .....((((-(((((((.(.(((((------------(....)------------)))))).))))))((((((((........).))))))).........)))))..... ( -21.20) >DroEre_CAF1 2286 105 + 1 UAUCUGCGA-UUAGAUAGAUACCUAUAAA------GAUAUUUAUACGAUUACCCCUGAGUUGUAUUUAUCGAUGGCAAACUAUCGUCCAUCGACAAUUGUUUAUCGCUAAAA .....((((-(.((((((...........------((((...((((((((.......)))))))).))))((((((........).))))).....)))))))))))..... ( -23.30) >DroYak_CAF1 2372 111 + 1 UAUCUGCGA-UUAGAUAGAUACCUAUACAUACGAGUAUAUUUAUACGAUUACCCCUGAGUUGUAUUUAUCGAUGGCAAACUAUCGUCCAUCGACAAUUGUUUAUCGCUAAAU .....((((-(.((((((.....(((((......)))))...((((((((.......))))))))...((((((((........).)))))))...)))))))))))..... ( -24.00) >DroPer_CAF1 2281 92 + 1 CAUCUGCAUACAAUAUAGAU--UG--AAAGAUGGG--CAUUAA-AC-----------GCAUGUAUUCUUCAAUGUUUCACUUUUAUAGUUCGCUCAUGGUUUAUCGCAUU-- (((((.....((((....))--))--..))))).(--(..(((-((-----------.((((...........((...(((.....)))..)).)))))))))..))...-- ( -12.70) >consensus UAUCUGCGA_UUAGAUAGAUACUUA_A_A________UAUUUA____________UGAGUUGUAUUUAUCGAUGGCAAACUUUCGUCCAUCGACAAUUGUUUAUCGCUAAAA .....((((....(((((((((.......................................)))))))))((((((........).)))))............))))..... ( -9.87 = -11.31 + 1.45)

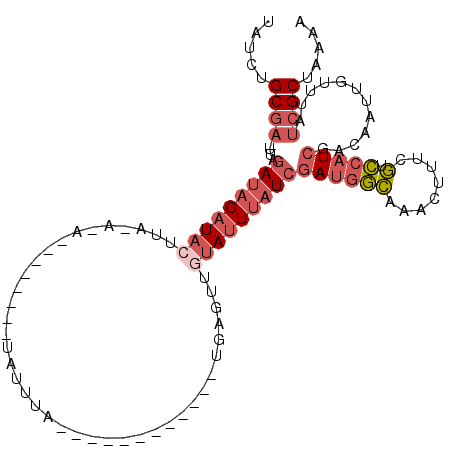
| Location | 11,369,516 – 11,369,609 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 71.14 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -13.37 |
| Energy contribution | -13.82 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
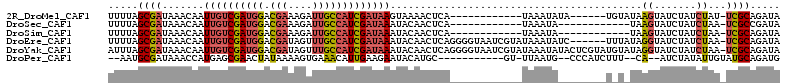
>2R_DroMel_CAF1 11369516 93 - 20766785 UUUUAGCGAUAAACAAUUGUCGAUGGACGAAAGAUUGCCAUCGAUAAGUAAAACUCA------------UAAAUAUA------UGUAUAAGUAUCUAUCUAU-UCGCAGAUA .....((((.......((((((((((.(....)....)))))))))).....(((.(------------((......------..))).)))..........-))))..... ( -21.00) >DroSec_CAF1 2271 87 - 1 UUUUAGCGAUAAACAAUUGUCGAUGGACGAAAGAUUGCCAUCGAUAAAUACAACUCA------------UAAAUA------------UAAGUAUCUAUCUAA-UCGCCGAUA ..(..(((((......((((((((((.(....)....)))))))))).....(((.(------------(....)------------).))).........)-))))..).. ( -20.50) >DroSim_CAF1 2265 87 - 1 UUUUAGCGAUAAACAAUUGUCGAUGGACGAAAGAUUGCCAUCGAUAAAUACAACUCA------------UAAAUA------------UAAGUAUCUAUCUAA-UCGCAGAUA .....(((((......((((((((((.(....)....)))))))))).....(((.(------------(....)------------).))).........)-))))..... ( -21.20) >DroEre_CAF1 2286 105 - 1 UUUUAGCGAUAAACAAUUGUCGAUGGACGAUAGUUUGCCAUCGAUAAAUACAACUCAGGGGUAAUCGUAUAAAUAUC------UUUAUAGGUAUCUAUCUAA-UCGCAGAUA .....(((((......((((((((((.(((....)))))))))))))...........(((((..(.((((((....------)))))).)....))))).)-))))..... ( -27.60) >DroYak_CAF1 2372 111 - 1 AUUUAGCGAUAAACAAUUGUCGAUGGACGAUAGUUUGCCAUCGAUAAAUACAACUCAGGGGUAAUCGUAUAAAUAUACUCGUAUGUAUAGGUAUCUAUCUAA-UCGCAGAUA .....(((((......((((((((((.(((....)))))))))))))...........(((((...((((...(((((......))))).)))).))))).)-))))..... ( -30.00) >DroPer_CAF1 2281 92 - 1 --AAUGCGAUAAACCAUGAGCGAACUAUAAAAGUGAAACAUUGAAGAAUACAUGC-----------GU-UUAAUG--CCCAUCUUU--CA--AUCUAUAUUGUAUGCAGAUG --...(((.(((((((((.....(((.....)))....(......)....)))).-----------))-))).))--).(((((..--((--((....)))).....))))) ( -11.20) >consensus UUUUAGCGAUAAACAAUUGUCGAUGGACGAAAGAUUGCCAUCGAUAAAUACAACUCA____________UAAAUA________U_U_UAAGUAUCUAUCUAA_UCGCAGAUA .....((((.......((((((((((.(((....)))))))))))))..........................................((.......))...))))..... (-13.37 = -13.82 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:20 2006