| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,172,225 – 2,172,335 |
| Length | 110 |
| Max. P | 0.995882 |

| Location | 2,172,225 – 2,172,335 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.63 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -14.07 |
| Energy contribution | -14.74 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
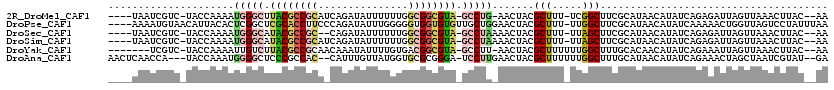
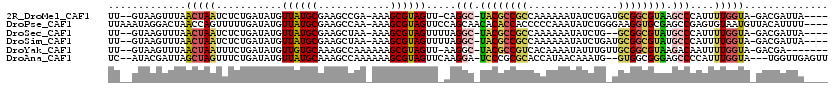
>2R_DroMel_CAF1 2172225 110 + 20766785 ----UAAUCGUC-UACCAAAAUGGGCUUACGCCGCAUCAGAUAUUUUUUGGCGGCGUA-GCCUG-AACUACGCUUU-UCGGCUUCGCAUAACAUAUCAGAGAUUAGUUAAACUUAC--AA ----(((((.((-(........(((((.(((((((..((((.....))))))))))))-)))).-......(((..-..)))...............))))))))...........--.. ( -28.80) >DroPse_CAF1 28901 115 + 1 ----AAAAUGUAACAUUACACUCGGCUCGCACCUUCCCAGAUAUUUGGGGGUGGUGUGUUGCUGGAACUACGCUUU-UUGGCUUCGCAUAACAUAUCAAAAACUGGUUAGUCCUAUUUAA ----.(((((..((.......(((((.(((((((((((((....))))))).))))))..)))))(((((.(.(((-((((..............))))))))))))).))..))))).. ( -29.24) >DroSec_CAF1 6756 109 + 1 ----UAAUCGUC-UACCAAAAUGGGCAUACGCCGC--CAGAUAUUUUUUGGCGGCGUA-GCCUAAAACUACGCUUU-UUAGCUUCGCAUAACAUAUCAGAGAUUAGUUAAACUUAC--AA ----(((((.((-(.......(((((.((((((((--((((.....))))))))))))-))))).......(((..-..)))...............))))))))...........--.. ( -36.80) >DroSim_CAF1 6813 111 + 1 ----UAAUCGUC-UACCAAAAUGGGCAUACGCCGCAUCAGAUAUUUUUUGGCGGCGUA-GCCUAAAACUACGCUUU-UUAGCUUCGCAUAACAUAUCAGAGAUUAGUUAAACUUAC--AA ----(((((.((-(.......(((((.((((((((..((((.....))))))))))))-))))).......(((..-..)))...............))))))))...........--.. ( -30.70) >DroYak_CAF1 7045 108 + 1 -------UCGUC-UACCAAAAUUGUCUUACGCCGCAACAAAUAUUUUGUGACGGCGUA-GCCUU-AACUACGCUUUUUUGGCUUUGCACAACAUAUCAGAAAUUAGUUAAACUUAC--AA -------.....-........((((.((((((((..((((.....))))..)))))))-)..((-(((((....(((((((...((.....))..))))))).)))))))....))--)) ( -22.00) >DroAna_CAF1 6882 112 + 1 AACUCAACCA---UACCAAAUGGGGCUCCCGCCAC--CAUUUGUUAUGGUGCGCGGGA-UCCUUGAACUACGCUUUUUUGGCUUUGCAUAACAUAUCAGAAACUAGCUAAUCGUAU--GA ........((---(((.....((((.(((((((((--(((.....)))))).))))))-.)))).......((((((((((...((.....))..)))))))..))).....))))--). ( -35.20) >consensus ____UAAUCGUC_UACCAAAAUGGGCUUACGCCGCA_CAGAUAUUUUGUGGCGGCGUA_GCCUA_AACUACGCUUU_UUGGCUUCGCAUAACAUAUCAGAAAUUAGUUAAACUUAC__AA .....................(((((.((((((((...............)))))))).))))).......(((.....)))...................................... (-14.07 = -14.74 + 0.67)
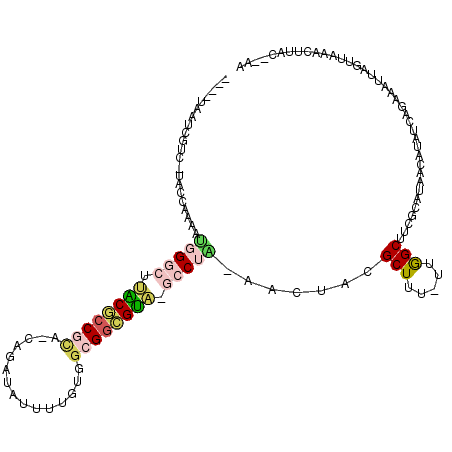
| Location | 2,172,225 – 2,172,335 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.63 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -16.27 |
| Energy contribution | -16.86 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
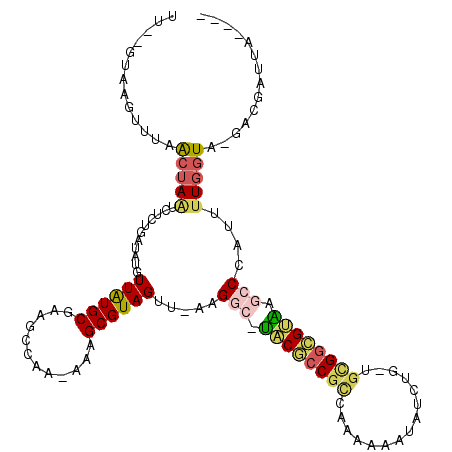
>2R_DroMel_CAF1 2172225 110 - 20766785 UU--GUAAGUUUAACUAAUCUCUGAUAUGUUAUGCGAAGCCGA-AAAGCGUAGUU-CAGGC-UACGCCGCCAAAAAAUAUCUGAUGCGGCGUAAGCCCAUUUUGGUA-GACGAUUA---- ..--....(((((.((((.....(((((.....(((.......-...(((((((.-...))-))))))))......))))).((((.(((....)))))))))))))-))).....---- ( -28.70) >DroPse_CAF1 28901 115 - 1 UUAAAUAGGACUAACCAGUUUUUGAUAUGUUAUGCGAAGCCAA-AAAGCGUAGUUCCAGCAACACACCACCCCCAAAUAUCUGGGAAGGUGCGAGCCGAGUGUAAUGUUACAUUUU---- .......((((((....((((((.....(((......)))..)-))))).))))))..((..(.((((...((((......))))..)))).).)).(((((((....))))))).---- ( -23.20) >DroSec_CAF1 6756 109 - 1 UU--GUAAGUUUAACUAAUCUCUGAUAUGUUAUGCGAAGCUAA-AAAGCGUAGUUUUAGGC-UACGCCGCCAAAAAAUAUCUG--GCGGCGUAUGCCCAUUUUGGUA-GACGAUUA---- ..--....(((((.((((...........((((((........-...)))))).....(((-((((((((((.........))--)))))))).)))....))))))-))).....---- ( -33.30) >DroSim_CAF1 6813 111 - 1 UU--GUAAGUUUAACUAAUCUCUGAUAUGUUAUGCGAAGCUAA-AAAGCGUAGUUUUAGGC-UACGCCGCCAAAAAAUAUCUGAUGCGGCGUAUGCCCAUUUUGGUA-GACGAUUA---- ..--....(((((.((((...........((((((........-...)))))).....(((-((((((((...............)))))))).)))....))))))-))).....---- ( -27.66) >DroYak_CAF1 7045 108 - 1 UU--GUAAGUUUAACUAAUUUCUGAUAUGUUGUGCAAAGCCAAAAAAGCGUAGUU-AAGGC-UACGCCGUCACAAAAUAUUUGUUGCGGCGUAAGACAAUUUUGGUA-GACGA------- ((--((..((.((((.............)))).))...(((((((....(((((.-...))-)))(((((.((((.....)))).))))).........))))))).-.))))------- ( -26.42) >DroAna_CAF1 6882 112 - 1 UC--AUACGAUUAGCUAGUUUCUGAUAUGUUAUGCAAAGCCAAAAAAGCGUAGUUCAAGGA-UCCCGCGCACCAUAACAAAUG--GUGGCGGGAGCCCCAUUUGGUA---UGGUUGAGUU ..--......(((((((....(..(.(((((((((............)))))).....((.-((((((.((((((.....)))--))))))))).)).))))..)..---)))))))... ( -33.70) >consensus UU__GUAAGUUUAACUAAUCUCUGAUAUGUUAUGCGAAGCCAA_AAAGCGUAGUU_AAGGC_UACGCCGCCAAAAAAUAUCUG_UGCGGCGUAAGCCCAUUUUGGUA_GACGAUUA____ .............(((((...........((((((............)))))).....(((.((((((((...............)))))))).)))....))))).............. (-16.27 = -16.86 + 0.59)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:49 2006