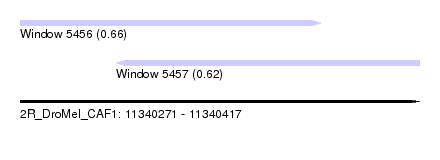
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,340,271 – 11,340,417 |
| Length | 146 |
| Max. P | 0.660911 |

| Location | 11,340,271 – 11,340,381 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -23.12 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
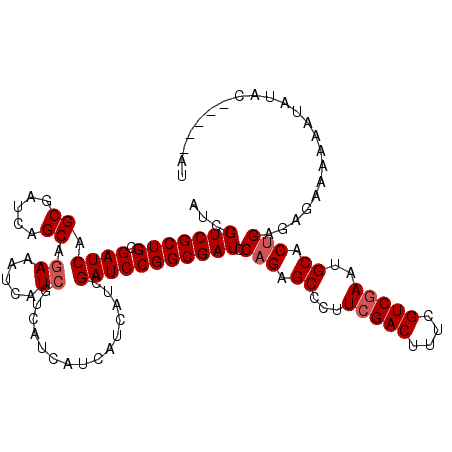
>2R_DroMel_CAF1 11340271 110 + 20766785 AUCGUCGCUGCGAUCAGCGAUCAGCAGAAAUCAUCGUCAUCAUCAUCAUCGAUCCGGCGAUCCAGAGCCCUUCGACUUUCGUCGAAUGCACUGAAAGAAAAUAUAUAC-----AU .(((((((((.((((.((.....)).((.....))...............))))))))))).(((.((..((((((....)))))).)).)))...))..........-----.. ( -28.70) >DroSec_CAF1 127995 110 + 1 AUCGUCGCUGCGAUCAGCGAUCAGCAGAAAUCAUCGUCAUCAUCAUCAUCGAUCCGGCGAUUCAGAGCUCUUCGACUUUCGUCGAAUGCACUGAGAGAAAUAAUAUAU-----AU .(((((((((.((((.((.....)).((.....))...............)))))))))))((((.((..((((((....)))))).)).))))))............-----.. ( -30.70) >DroSim_CAF1 133733 105 + 1 AUCGUCGCUGCGAUCAGCGAUCAGCAGAAAUCAUC---AUCAUCAUCAUCGAUCCGGCGAUUCAGAGCCCUUCGACUUUCGUCAAAUGCACCGAGAGAAAAAAU--AU-----AU ...(((((((....)))))))..............---..........((((...(((........)))..))))((((((..........)))))).......--..-----.. ( -22.00) >DroYak_CAF1 131096 115 + 1 AUCGUCGCUGCGAUCAGCGAUCAGCAAAAAUCAUCGUCAUCAUCAUCAUCGAUCCGGCGAUCCAGAGCCCUUCGACUUUCGUCGAUUGCACUGAGAAAAAAUGUAUACUAUGUUU ...(((((((....)))))))..(((.......(((((...(((......)))..)))))(((((.((...(((((....)))))..)).))).)).....)))........... ( -28.10) >consensus AUCGUCGCUGCGAUCAGCGAUCAGCAGAAAUCAUCGUCAUCAUCAUCAUCGAUCCGGCGAUCCAGAGCCCUUCGACUUUCGUCGAAUGCACUGAGAGAAAAAAUAUAC_____AU ...(((((((.((((.((.....)).((.....))...............))))))))))).(((.((...(((((....)))))..)).)))...................... (-23.12 = -23.88 + 0.75)

| Location | 11,340,306 – 11,340,417 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.82 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -19.80 |
| Energy contribution | -19.43 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
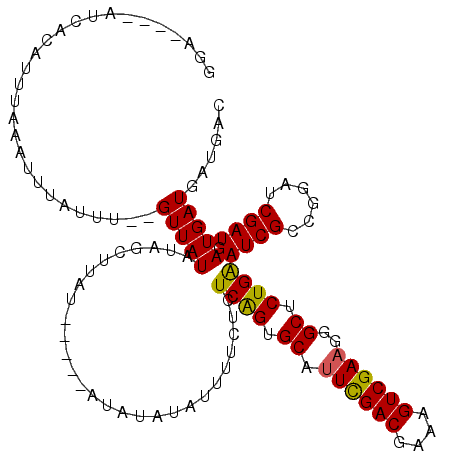
>2R_DroMel_CAF1 11340306 111 - 20766785 GGA----AUCUCAUUUAAAUUUUUUUGUGUUAUAUAGCUUAU-----GUAUAUAUUUUCUUUCAGUGCAUUCGACGAAAGUCGAAGGGCUCUGGAUCGCCGGAUCGAUGAUGAUGAUGAC ...----.(((((((...(((..((((.((((((((.....)-----)))))........(((((.((.((((((....))))))..)).)))))..))))))..)))...))))).)). ( -29.10) >DroSec_CAF1 128030 109 - 1 GGA----AUCACAUUUAAAUUUAUUU--GUUAUAUAGCUUAU-----AUAUAUUAUUUCUCUCAGUGCAUUCGACGAAAGUCGAAGAGCUCUGAAUCGCCGGAUCGAUGAUGAUGAUGAC ...----.(((((((...(((.((((--(.((((((.....)-----))))).........((((.((.((((((....))))))..)).)))).....))))).)))...)))).))). ( -28.10) >DroSim_CAF1 133768 104 - 1 GGA----AUCACAUUUAAAUUCAUUU--GUUAUAUAGCUUAU-----AU--AUUUUUUCUCUCGGUGCAUUUGACGAAAGUCGAAGGGCUCUGAAUCGCCGGAUCGAUGAUGAUGAU--- ...----((((((((...((((....--..((((((...)))-----))--).........((((.((.((((((....))))))..)).))))......)))).)))).))))...--- ( -23.00) >DroYak_CAF1 131131 118 - 1 GGAAUGCAUCCCAAUUCAAUUUCUUU--GUUAUAUAUACAAAACAUAGUAUACAUUUUUUCUCAGUGCAAUCGACGAAAGUCGAAGGGCUCUGGAUCGCCGGAUCGAUGAUGAUGAUGAC .((((........)))).......(.--.((((.(((((........)))))((((...((.(((.((..(((((....)))))...)).)))((((....)))))).))))))))..). ( -28.80) >consensus GGA____AUCACAUUUAAAUUUAUUU__GUUAUAUAGCUUAU_____AUAUAUAUUUUCUCUCAGUGCAUUCGACGAAAGUCGAAGGGCUCUGAAUCGCCGGAUCGAUGAUGAUGAUGAC ............................(((((............................((((.((.((((((....))))))..)).))))((((......)))).)))))...... (-19.80 = -19.43 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:02 2006