
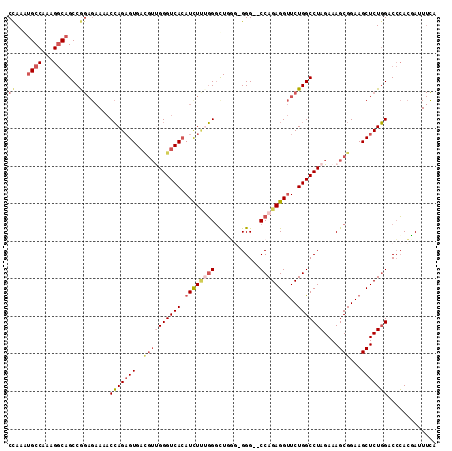
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,339,040 – 11,339,293 |
| Length | 253 |
| Max. P | 0.979674 |

| Location | 11,339,040 – 11,339,154 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -43.42 |
| Consensus MFE | -26.82 |
| Energy contribution | -28.45 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11339040 114 + 20766785 CCAAAUGCCAAAGGCAGCCGGAGAAAACCAGAGUGACGUUGGGUCACAUCUUUGGGUUGGGGGGGGUCCAGAGGUUCUGGCCUAGAAAGCGGAAGCUCUGGACCCACUAUUUCA ((((...(((((((.....((......))...(((((.....))))).))))))).))))((.((((((((((.(((((..........))))).)))))))))).))...... ( -47.80) >DroSec_CAF1 126783 112 + 1 CCAAAUGCCAAAGGCAGCCGGAGAAAACCAGAGUGACGUUGGGUCACAUCUUUGGGUUGGGGGGG--CCAGAGGUUCUGGCCUAGAAAGCGGAAGCUCUGGACCCACUAUUUCA ((...((((...))))...)).((((...((((((((.....))))).))).((((((.((.(((--((((.....)))))))....(((....))))).))))))...)))). ( -45.70) >DroSim_CAF1 132517 112 + 1 CCAAAUGCCAAAGGCAGCCGGAGAAAACCAGAGUGACGUUGGGUCACAUCUUGGGGUUGGAGGGG--CCAGAGGUUCUGGCCUAGAAAGCGGAAGCUCUGGACCCACUAUUUCA ((...((((...))))...)).((((.((((((((((.....))))).)).)))...(((.(((.--((((((.(((((..........))))).)))))).))).))))))). ( -43.00) >DroEre_CAF1 130691 110 + 1 CCAAAUGCCAAAGGCAGCCAGAGAAAACCAGAGUGAUGUUGGGUCACAUCUCUGGGCUCGG-GGG--CCAGAGGUUCUGGCCUAGAAAGCGGAAGCUCUGGACCCACGA-UUCA ((...((((...))))....(((....((((((((.((......)))).)))))).)))))-(((--((((((.(((((..........))))).)))))).)))....-.... ( -42.70) >DroYak_CAF1 129840 111 + 1 CUAAAUGCCAAAGGCAGCCAGAGAAAACCAGAGUGACGUCGGGUCACAUUUCUGGGCUCAG-GGG--CCAGAGGUUCUGGCCUAGAAAGCGGAAGCUCUGGACCCACGAAUUUC .....((((...)))).((((((....((((((((((.....)))))...)))))(((...-(((--((((.....)))))))....))).....))))))............. ( -43.60) >DroAna_CAF1 127815 88 + 1 CCAAGAGC------CAGCCG-------CGAGUGUG----CGGGUCAAAUCUGGUGUCUUU--GGG--CCAGAGGCUCUGGCCUAGAAAGCGGAAGCUCUUGUCGGAGGA----- .(((((((------...(((-------(....(..----(.((......)).)..).(((--(((--((((.....))))))))))..))))..)))))))........----- ( -37.70) >consensus CCAAAUGCCAAAGGCAGCCGGAGAAAACCAGAGUGACGUUGGGUCACAUCUUUGGGCUGGG_GGG__CCAGAGGUUCUGGCCUAGAAAGCGGAAGCUCUGGACCCACGAUUUCA ((...((((...))))...))......(((((((..((((((((((.((((((((............))))))))..)))))))....)))...)))))))............. (-26.82 = -28.45 + 1.63)


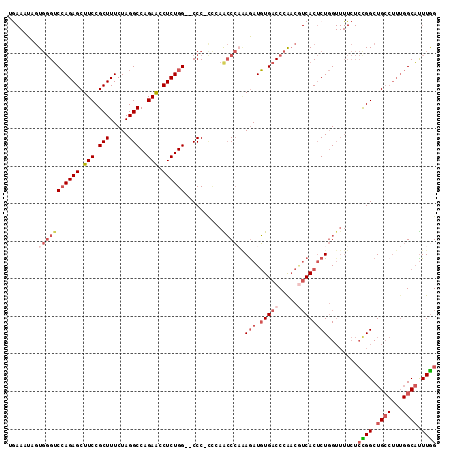
| Location | 11,339,040 – 11,339,154 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -20.99 |
| Energy contribution | -23.03 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11339040 114 - 20766785 UGAAAUAGUGGGUCCAGAGCUUCCGCUUUCUAGGCCAGAACCUCUGGACCCCCCCCAACCCAAAGAUGUGACCCAACGUCACUCUGGUUUUCUCCGGCUGCCUUUGGCAUUUGG .........((((((((((.(((.(((.....)))..))).))))))))))...((((.((((((..(((((.....))))).((((......))))....))))))...)))) ( -42.60) >DroSec_CAF1 126783 112 - 1 UGAAAUAGUGGGUCCAGAGCUUCCGCUUUCUAGGCCAGAACCUCUGG--CCCCCCCAACCCAAAGAUGUGACCCAACGUCACUCUGGUUUUCUCCGGCUGCCUUUGGCAUUUGG .........(((.((((((.(((.(((.....)))..))).))))))--)))..((((.((((((..(((((.....))))).((((......))))....))))))...)))) ( -37.60) >DroSim_CAF1 132517 112 - 1 UGAAAUAGUGGGUCCAGAGCUUCCGCUUUCUAGGCCAGAACCUCUGG--CCCCUCCAACCCCAAGAUGUGACCCAACGUCACUCUGGUUUUCUCCGGCUGCCUUUGGCAUUUGG .((((.((.(((.((((((.(((.(((.....)))..))).))))))--)))))......(((.((.(((((.....)))))))))).)))).((((.((((...)))).)))) ( -37.40) >DroEre_CAF1 130691 110 - 1 UGAA-UCGUGGGUCCAGAGCUUCCGCUUUCUAGGCCAGAACCUCUGG--CCC-CCGAGCCCAGAGAUGUGACCCAACAUCACUCUGGUUUUCUCUGGCUGCCUUUGGCAUUUGG ....-(((.((((((((((.....((((....((((((.....))))--)).-..))))((((((((((......)))))..)))))....))))))..)))).)))....... ( -40.50) >DroYak_CAF1 129840 111 - 1 GAAAUUCGUGGGUCCAGAGCUUCCGCUUUCUAGGCCAGAACCUCUGG--CCC-CUGAGCCCAGAAAUGUGACCCGACGUCACUCUGGUUUUCUCUGGCUGCCUUUGGCAUUUAG .(((((((.((((((((((.....((((....((((((.....))))--)).-..))))(((((...(((((.....))))))))))....))))))..)))).))).)))).. ( -41.90) >DroAna_CAF1 127815 88 - 1 -----UCCUCCGACAAGAGCUUCCGCUUUCUAGGCCAGAGCCUCUGG--CCC--AAAGACACCAGAUUUGACCCG----CACACUCG-------CGGCUG------GCUCUUGG -----........((((((((.((((......((((((.....))))--))(--(((.........)))).....----.......)-------)))..)------))))))). ( -32.50) >consensus UGAAAUAGUGGGUCCAGAGCUUCCGCUUUCUAGGCCAGAACCUCUGG__CCC_CCCAACCCAAAGAUGUGACCCAACGUCACUCUGGUUUUCUCCGGCUGCCUUUGGCAUUUGG ........(((((((((((.(((.(((.....)))..))).))))))..........))))).(((.(((((.....))))))))........((((.((((...)))).)))) (-20.99 = -23.03 + 2.04)

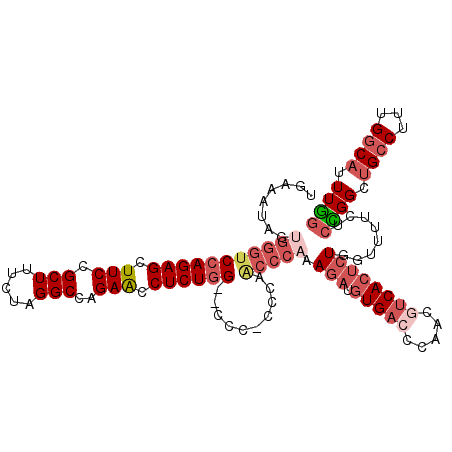
| Location | 11,339,080 – 11,339,194 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -42.08 |
| Consensus MFE | -32.30 |
| Energy contribution | -32.46 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11339080 114 + 20766785 GGGUCACAUCUUUGGGUUGGGGGGGGUCCAGAGGUUCUGGCCUAGAAAGCGGAAGCUCUGGACCCACUAUUUCAGGCUAAUUGGGAAAGAGGAAGUCAGGUGACAACGGAAGAU ..(((((.(((((((.(((..(.((((((((((.(((((..........))))).))))))))))....)..))).))))...)))..((.....))..)))))..(....).. ( -44.00) >DroSec_CAF1 126823 112 + 1 GGGUCACAUCUUUGGGUUGGGGGGG--CCAGAGGUUCUGGCCUAGAAAGCGGAAGCUCUGGACCCACUAUUUCAGGCUAAUUGAGAAAGAGGAAGUCAGGCGACAACGGAAGAU ..(((.(..(((((((((.((.(((--((((.....)))))))....(((....))))).))))).((.((((((.....)))))).))..))))....).)))..(....).. ( -40.80) >DroSim_CAF1 132557 112 + 1 GGGUCACAUCUUGGGGUUGGAGGGG--CCAGAGGUUCUGGCCUAGAAAGCGGAAGCUCUGGACCCACUAUUUCAGGCUAAUUGGGAAAGAGGAAGUCAGGUGACAACGGAAGAU ..(((((..((((((.(..((.(((--((((.....))))))).....((....))))..).))).((.(..(((.....)))..).))...)))....)))))..(....).. ( -39.80) >DroEre_CAF1 130731 110 + 1 GGGUCACAUCUCUGGGCUCGG-GGG--CCAGAGGUUCUGGCCUAGAAAGCGGAAGCUCUGGACCCACGA-UUCAGGCUAAUUGGGAAAGAGGAAGUCAGGUGACAACGGAAGAU ..(((((...((((((..(((-(((--((((.....)))))))....(((....))))))..)))).))-....((((..((......))...))))..)))))..(....).. ( -40.80) >DroYak_CAF1 129880 111 + 1 GGGUCACAUUUCUGGGCUCAG-GGG--CCAGAGGUUCUGGCCUAGAAAGCGGAAGCUCUGGACCCACGAAUUUCGGCUAAUUGGUAAAGAGGAAGUCGGGUGACAACGGAAGAU ..(((((..(((((((..(((-(((--((((.....)))))))....(((....))))))..)))).)))..((((((..((......))...)))))))))))..(....).. ( -45.00) >consensus GGGUCACAUCUUUGGGUUGGGGGGG__CCAGAGGUUCUGGCCUAGAAAGCGGAAGCUCUGGACCCACUAUUUCAGGCUAAUUGGGAAAGAGGAAGUCAGGUGACAACGGAAGAU ..(((((......(((.(((.(((...((((((.(((((..........))))).)))))).))).))).))).((((..((......))...))))..)))))..(....).. (-32.30 = -32.46 + 0.16)


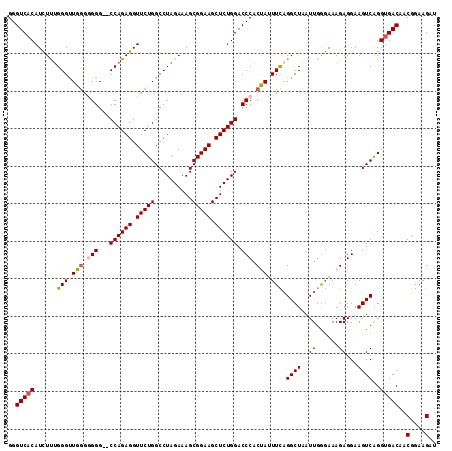
| Location | 11,339,080 – 11,339,194 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -22.28 |
| Energy contribution | -22.08 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11339080 114 - 20766785 AUCUUCCGUUGUCACCUGACUUCCUCUUUCCCAAUUAGCCUGAAAUAGUGGGUCCAGAGCUUCCGCUUUCUAGGCCAGAACCUCUGGACCCCCCCCAACCCAAAGAUGUGACCC ..........(((((........((.((((...........)))).)).((((((((((.(((.(((.....)))..))).))))))))))................))))).. ( -30.10) >DroSec_CAF1 126823 112 - 1 AUCUUCCGUUGUCGCCUGACUUCCUCUUUCUCAAUUAGCCUGAAAUAGUGGGUCCAGAGCUUCCGCUUUCUAGGCCAGAACCUCUGG--CCCCCCCAACCCAAAGAUGUGACCC (((((..((((..((((((((..((.((((...........)))).))..))).))).))............((((((.....))))--))....))))...)))))....... ( -29.30) >DroSim_CAF1 132557 112 - 1 AUCUUCCGUUGUCACCUGACUUCCUCUUUCCCAAUUAGCCUGAAAUAGUGGGUCCAGAGCUUCCGCUUUCUAGGCCAGAACCUCUGG--CCCCUCCAACCCCAAGAUGUGACCC ..........(((((..((((..((.((((...........)))).))..)))).(((((....)))))...((((((.....))))--))................))))).. ( -27.50) >DroEre_CAF1 130731 110 - 1 AUCUUCCGUUGUCACCUGACUUCCUCUUUCCCAAUUAGCCUGAA-UCGUGGGUCCAGAGCUUCCGCUUUCUAGGCCAGAACCUCUGG--CCC-CCGAGCCCAGAGAUGUGACCC ..........(((((((((...............)))).(((..-(((.(((.((((((.(((.(((.....)))..))).))))))--.))-))))...)))....))))).. ( -30.86) >DroYak_CAF1 129880 111 - 1 AUCUUCCGUUGUCACCCGACUUCCUCUUUACCAAUUAGCCGAAAUUCGUGGGUCCAGAGCUUCCGCUUUCUAGGCCAGAACCUCUGG--CCC-CUGAGCCCAGAAAUGUGACCC ..........(((((.((.((...............)).))...(((.((((..((((((....))).....((((((.....))))--)).-)))..)))))))..))))).. ( -32.16) >consensus AUCUUCCGUUGUCACCUGACUUCCUCUUUCCCAAUUAGCCUGAAAUAGUGGGUCCAGAGCUUCCGCUUUCUAGGCCAGAACCUCUGG__CCCCCCCAACCCAAAGAUGUGACCC ..........(((((..((.....))......................(((((((((((.(((.(((.....)))..))).))))))..........))))).....))))).. (-22.28 = -22.08 + -0.20)

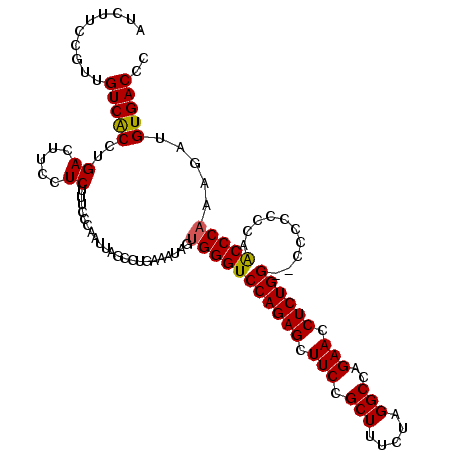
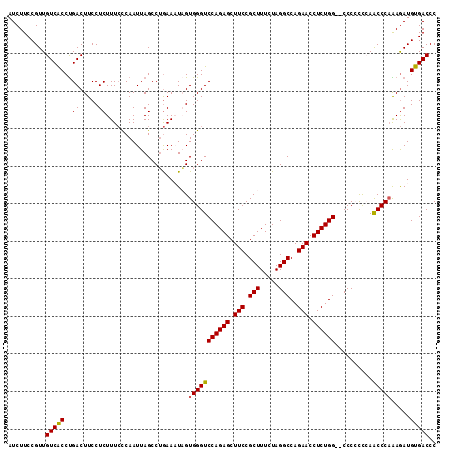
| Location | 11,339,154 – 11,339,270 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -26.45 |
| Energy contribution | -26.07 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11339154 116 + 20766785 GGCUAAUUGGGAAAGAGGAAGUCAGGUGACAACGGAAGAUGUGGCUUUAUAAAGCUGAGGAACAGUUUCAGCCGACUCCUCUACAGCUUCAUUAGAAGUAGCUAUUAAGGUCUUA----G ((((...(((...((((((.(((((((.(((.(....).))).))))......(((((((.....))))))).)))))))))...(((((....)))))..)))....))))...----. ( -36.40) >DroSec_CAF1 126895 116 + 1 GGCUAAUUGAGAAAGAGGAAGUCAGGCGACAACGGAAGAUGUGGCUUCAUAAAGCUGAGGAACAGUCUCAGCCCGCUCCUCUACAACUUCAUUAGAAGUAGCUAUUAAGGUCUUA----A ((((.........((((((.((.((((.(((.(....).))).))))......(((((((.....)))))))..))))))))...(((((....)))))))))............----. ( -35.60) >DroSim_CAF1 132629 116 + 1 GGCUAAUUGGGAAAGAGGAAGUCAGGUGACAACGGAAGAUGGGGCUUCAUAAAGCUGAGGAACAGUUUUAGCCCACUCCUCUACAGCUUCAAUAGAAGUAGCUAAUAAGGUCUUA----A ((((.(((((...((((((.(((....)))..(....)...(((((....(((((((.....))))))))))))..))))))...(((((....)))))..)))))..))))...----. ( -35.60) >DroYak_CAF1 129951 107 + 1 GGCUAAUUGGUAAAGAGGAAGUCGGGUGACAACGGAAGAUGGGGCUUCGUAAAACUUAGGAACAGUUUGAG-----UUGUCUACAGCUUCAUUAGAAGCUGCUAUU--------AGAAAC ..(((((.(((......((((((.(....)..(....)....))))))(((.((((((((.....))))))-----))...)))((((((....))))))))))))--------)).... ( -27.90) >consensus GGCUAAUUGGGAAAGAGGAAGUCAGGUGACAACGGAAGAUGGGGCUUCAUAAAGCUGAGGAACAGUUUCAGCCCACUCCUCUACAGCUUCAUUAGAAGUAGCUAUUAAGGUCUUA____A ((((.........((((((....((((.(((.(....).))).))))......(((((((.....)))))))....))))))...(((((....)))))))))................. (-26.45 = -26.07 + -0.38)

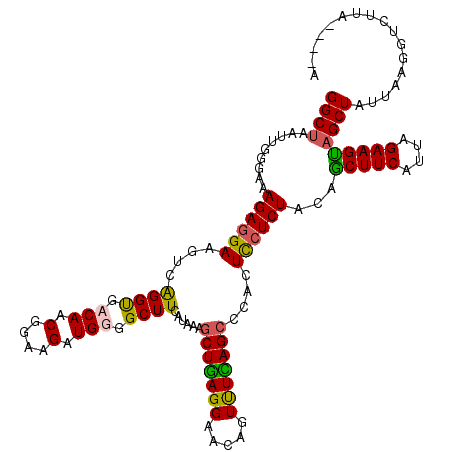
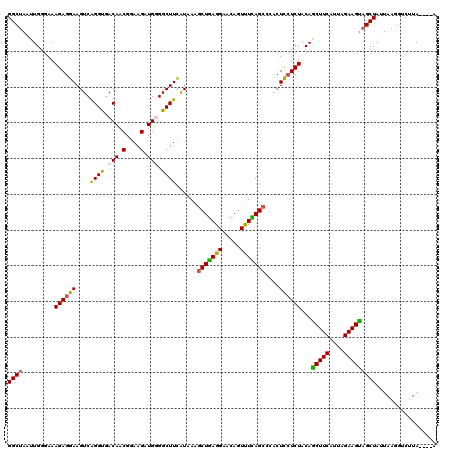
| Location | 11,339,154 – 11,339,270 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -16.49 |
| Energy contribution | -17.68 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11339154 116 - 20766785 C----UAAGACCUUAAUAGCUACUUCUAAUGAAGCUGUAGAGGAGUCGGCUGAAACUGUUCCUCAGCUUUAUAAAGCCACAUCUUCCGUUGUCACCUGACUUCCUCUUUCCCAAUUAGCC (----(((.......((((((.(.......).))))))(((((((((((((.((((((.....))).)))....))))(((........))).....))).)))))).......)))).. ( -26.60) >DroSec_CAF1 126895 116 - 1 U----UAAGACCUUAAUAGCUACUUCUAAUGAAGUUGUAGAGGAGCGGGCUGAGACUGUUCCUCAGCUUUAUGAAGCCACAUCUUCCGUUGUCGCCUGACUUCCUCUUUCUCAAUUAGCC .----.............((((((((....))))(((.((((((((((((((((.......))))))........((.(((........))).))))).).))))))....))).)))). ( -29.70) >DroSim_CAF1 132629 116 - 1 U----UAAGACCUUAUUAGCUACUUCUAUUGAAGCUGUAGAGGAGUGGGCUAAAACUGUUCCUCAGCUUUAUGAAGCCCCAUCUUCCGUUGUCACCUGACUUCCUCUUUCCCAAUUAGCC .----.............((((((((...(((((((...((((((..(.......)..))))))))))))).))))..............(((....)))...............)))). ( -27.20) >DroYak_CAF1 129951 107 - 1 GUUUCU--------AAUAGCAGCUUCUAAUGAAGCUGUAGACAA-----CUCAAACUGUUCCUAAGUUUUACGAAGCCCCAUCUUCCGUUGUCACCCGACUUCCUCUUUACCAAUUAGCC ....((--------(((.((((((((....)))))))).(((((-----(..(((((.......)))))...((((......)))).))))))....................))))).. ( -25.90) >consensus U____UAAGACCUUAAUAGCUACUUCUAAUGAAGCUGUAGAGGAGUGGGCUGAAACUGUUCCUCAGCUUUAUGAAGCCACAUCUUCCGUUGUCACCUGACUUCCUCUUUCCCAAUUAGCC ..................((((((((...((((((((..(((.(((........))).)))..)))))))).))))..............(((....)))...............)))). (-16.49 = -17.68 + 1.19)

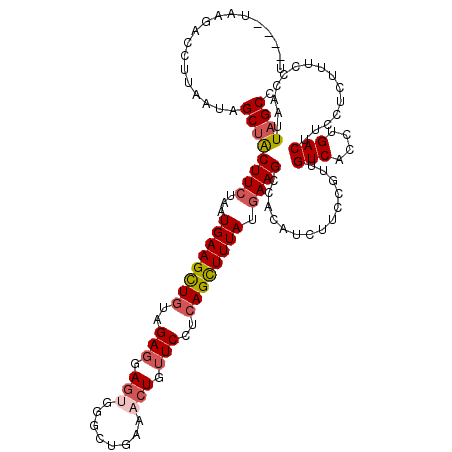
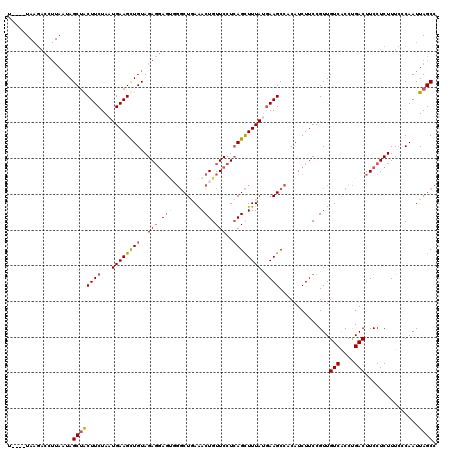
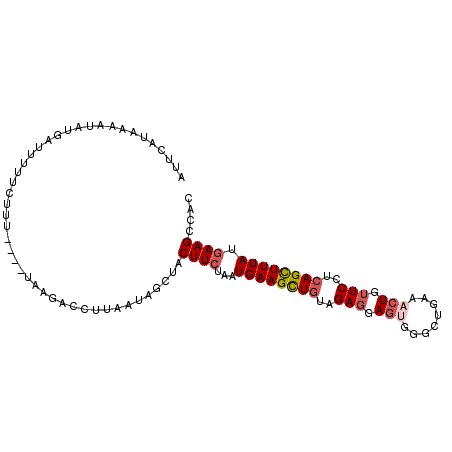
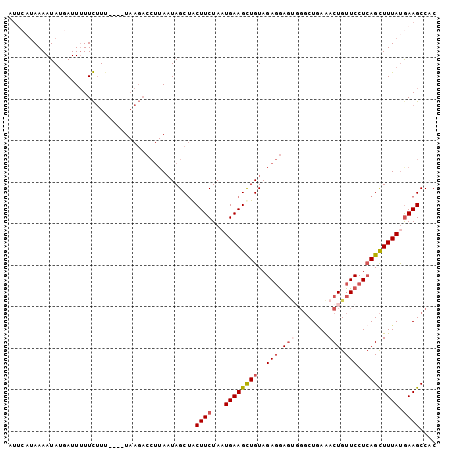
| Location | 11,339,194 – 11,339,293 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.57 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -12.25 |
| Energy contribution | -13.38 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11339194 99 - 20766785 AUUCAUAAAAUAUGAUUUUUCUUC----UAAGACCUUAAUAGCUACUUCUAAUGAAGCUGUAGAGGAGUCGGCUGAAACUGUUCCUCAGCUUUAUAAAGCCAC ..(((((...))))).........----.....((((.((((((.(.......).)))))).))))....((((.((((((.....))).)))....)))).. ( -21.20) >DroSec_CAF1 126935 99 - 1 AUUCAUAAAAUACGAUUUUUCUUU----UAAGACCUUAAUAGCUACUUCUAAUGAAGUUGUAGAGGAGCGGGCUGAGACUGUUCCUCAGCUUUAUGAAGCCAC .((((((((....((....))...----.............((((((((....)))))....(((((((((.......))))))))))))))))))))..... ( -27.20) >DroSim_CAF1 132669 93 - 1 ------AAAAUAUGAUUUUUCUUU----UAAGACCUUAUUAGCUACUUCUAUUGAAGCUGUAGAGGAGUGGGCUAAAACUGUUCCUCAGCUUUAUGAAGCCCC ------..................----.................((((...(((((((...((((((..(.......)..))))))))))))).)))).... ( -21.80) >DroYak_CAF1 129991 90 - 1 UUACAUAAAAUAUAAUUAUUUAUGUUUCU--------AAUAGCAGCUUCUAAUGAAGCUGUAGACAA-----CUCAAACUGUUCCUAAGUUUUACGAAGCCCC .......(((((....)))))..(((((.--------....((((((((....))))))))......-----...(((((.......)))))...)))))... ( -18.20) >consensus AUUCAUAAAAUAUGAUUUUUCUUU____UAAGACCUUAAUAGCUACUUCUAAUGAAGCUGUAGAGGAGUGGGCUGAAACUGUUCCUCAGCUUUAUGAAGCCAC .............................................((((...((((((((..(((.(((........))).)))..)))))))).)))).... (-12.25 = -13.38 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:59 2006