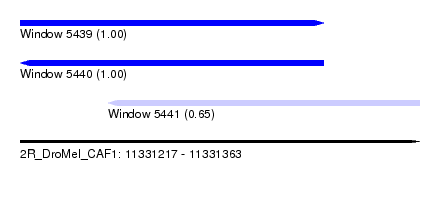
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,331,217 – 11,331,363 |
| Length | 146 |
| Max. P | 0.999951 |

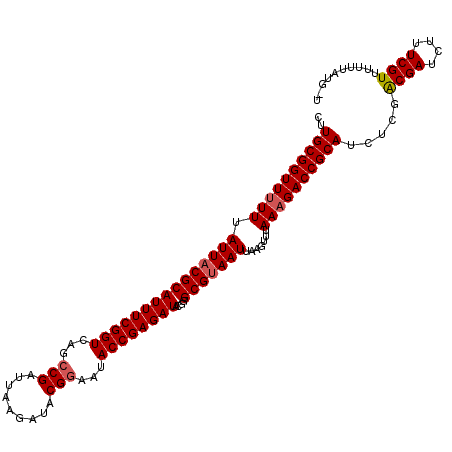
| Location | 11,331,217 – 11,331,328 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 93.09 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -31.34 |
| Energy contribution | -31.78 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.80 |
| SVM RNA-class probability | 0.999951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11331217 111 + 20766785 CUUGCGGUUUUUUAUUACGCAUUUCGGUCAGGCGAUUAAGAUACGAAAUACCGAGAUACGGGCGUAAUUAAGUUUAAAGACCGCAUCUCGGCGAUCUUUCGUUU-UUAUGUU ..((((((((((.((((((((((((((((..........))).)))))).(((.....)))))))))).......))))))))))....(((((....))))).-....... ( -31.40) >DroSec_CAF1 118621 112 + 1 CUUGCGGUUUUUUAUUACGCAUUUCGGUGAGCCGAUUAAGAUACGGAAUACCGAGAUACGGGCGUAAUUAAGUUUAAAGACCGCAUCUCGACGAUCUUUCGUUUUUUAUGUU ..((((((((((.((((((((((((((((..(((.........)))..)))))))))....))))))).......))))))))))....(((((....)))))......... ( -37.00) >DroSim_CAF1 122466 112 + 1 CUUGCGGUUUUUUAUUACGCAUUUCGGUCAGCCGAUUAAGAUACGGAAUACCGAGAUACGGGCGUAAUUAAGUUUAAAGACCGCAUCUCGACGAUCUUUCGUUUUUUAUGAU ..((((((((((.(((((((((((((((...(((.........)))...))))))))....))))))).......))))))))))....(((((....)))))......... ( -35.10) >DroEre_CAF1 119912 104 + 1 CUUGCGGUU-UUUAUUACGCAUUUCGGUCAGCCGAUUAAGAUACGGGAUACCGAGAUACGGGCGUAAUUAAGUUUAAAGACCGCAUCUCGACGAUCUUUCGU-UUU------ ..(((((((-((((((((((((((((((...(((.........)))...))))))))....)))))).......)))))))))))....(((((....))))-)..------ ( -33.91) >DroYak_CAF1 121494 106 + 1 CUUGCGGUUUUUUAUUACGCAUUUCGGUCAGCCGAUUAAGAUACGGAAUACCGAGAUACGGGCGCAAUUAAGUUUAAAGACCGCAUCUCGACGAUCUUUCGUGUUU------ ..((((((((((.....((.((((((((...(((.........)))...)))))))).))(((........))).)))))))))).....((((....))))....------ ( -30.50) >consensus CUUGCGGUUUUUUAUUACGCAUUUCGGUCAGCCGAUUAAGAUACGGAAUACCGAGAUACGGGCGUAAUUAAGUUUAAAGACCGCAUCUCGACGAUCUUUCGUUUUUUAUG_U ..((((((((((.(((((((((((((((...(((.........)))...))))))))....))))))).......)))))))))).....((((....)))).......... (-31.34 = -31.78 + 0.44)

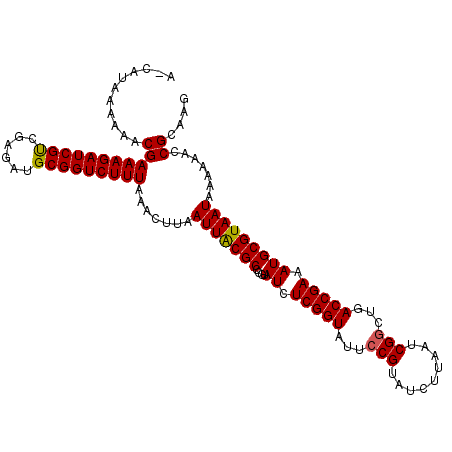
| Location | 11,331,217 – 11,331,328 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 93.09 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -25.40 |
| Energy contribution | -25.28 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11331217 111 - 20766785 AACAUAA-AAACGAAAGAUCGCCGAGAUGCGGUCUUUAAACUUAAUUACGCCCGUAUCUCGGUAUUUCGUAUCUUAAUCGCCUGACCGAAAUGCGUAAUAAAAAACCGCAAG .......-..((((((....((((((((((((.(...............).)))))))))))).)))))).......(((......)))..((((...........)))).. ( -28.76) >DroSec_CAF1 118621 112 - 1 AACAUAAAAAACGAAAGAUCGUCGAGAUGCGGUCUUUAAACUUAAUUACGCCCGUAUCUCGGUAUUCCGUAUCUUAAUCGGCUCACCGAAAUGCGUAAUAAAAAACCGCAAG ..............((((((((((((((((((.(...............).))))))))))).....)).)))))..((((....))))..((((...........)))).. ( -25.76) >DroSim_CAF1 122466 112 - 1 AUCAUAAAAAACGAAAGAUCGUCGAGAUGCGGUCUUUAAACUUAAUUACGCCCGUAUCUCGGUAUUCCGUAUCUUAAUCGGCUGACCGAAAUGCGUAAUAAAAAACCGCAAG ...........(((((((((((......))))))))).......(((((((....((.(((((...(((.........)))...))))).))))))))).......)).... ( -26.00) >DroEre_CAF1 119912 104 - 1 ------AAA-ACGAAAGAUCGUCGAGAUGCGGUCUUUAAACUUAAUUACGCCCGUAUCUCGGUAUCCCGUAUCUUAAUCGGCUGACCGAAAUGCGUAAUAAA-AACCGCAAG ------...-((((....)))).....((((((.((((.......((((((....((.(((((...(((.........)))...))))).))))))))))))-.)))))).. ( -26.41) >DroYak_CAF1 121494 106 - 1 ------AAACACGAAAGAUCGUCGAGAUGCGGUCUUUAAACUUAAUUGCGCCCGUAUCUCGGUAUUCCGUAUCUUAAUCGGCUGACCGAAAUGCGUAAUAAAAAACCGCAAG ------.......(((((((((......)))))))))........(((((..(((((.(((((...(((.........)))...))))).)))))...........))))). ( -27.72) >consensus A_CAUAAAAAACGAAAGAUCGUCGAGAUGCGGUCUUUAAACUUAAUUACGCCCGUAUCUCGGUAUUCCGUAUCUUAAUCGGCUGACCGAAAUGCGUAAUAAAAAACCGCAAG ...........(((((((((((......))))))))).......(((((((....((.(((((...(((.........)))...))))).))))))))).......)).... (-25.40 = -25.28 + -0.12)

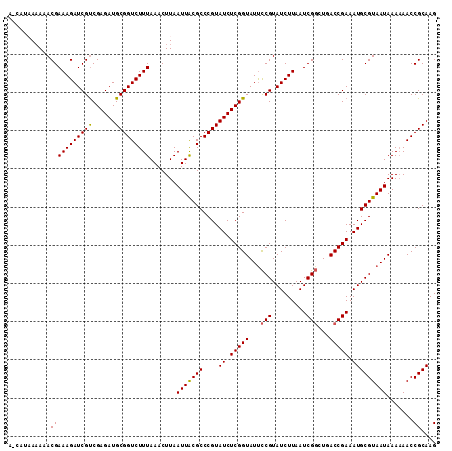
| Location | 11,331,249 – 11,331,363 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.04 |
| Mean single sequence MFE | -24.49 |
| Consensus MFE | -20.98 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11331249 114 - 20766785 AUUGUGUAACUAACCGUUGUCACCGUUGUCAUUAAAACAUAA-AAACGAAAGAUCGCCGAGAUGCGGUCUUUAAACUUAAUUACGCCCGUAUCUCGGUAUUUCGUAUCUUAAUCG ...(((((((.....)))).))).(((........)))....-..((((((....((((((((((((.(...............).)))))))))))).)))))).......... ( -29.76) >DroSec_CAF1 118653 106 - 1 AUUGUGUAACUAACCGU---------UGUCAUCAAAACAUAAAAAACGAAAGAUCGUCGAGAUGCGGUCUUUAAACUUAAUUACGCCCGUAUCUCGGUAUUCCGUAUCUUAAUCG .(((((((((.....))---------)).)).)))...........(((((((((((((((((((((.(...............).))))))))))).....)).)))))..))) ( -22.36) >DroSim_CAF1 122498 115 - 1 AUUGUGUAACUAACCGUUGUCACCGUUGUCAUCAAAUCAUAAAAAACGAAAGAUCGUCGAGAUGCGGUCUUUAAACUUAAUUACGCCCGUAUCUCGGUAUUCCGUAUCUUAAUCG ...(((((((.....)))).)))((((.................)))).((((((((((((((((((.(...............).))))))))))).....)).)))))..... ( -23.59) >DroEre_CAF1 119943 105 - 1 AUUGUGUAACUAACCGUUGUCACCGU---CAUUAA------AAA-ACGAAAGAUCGUCGAGAUGCGGUCUUUAAACUUAAUUACGCCCGUAUCUCGGUAUCCCGUAUCUUAAUCG ...(((((((.....)))).)))...---......------...-.(((((((((((((((((((((.(...............).))))))))))).....)).)))))..))) ( -23.36) >DroYak_CAF1 121526 106 - 1 AUUGUGUAACUAACCGUUGUCACCGU---CAUUAA------AAACACGAAAGAUCGUCGAGAUGCGGUCUUUAAACUUAAUUGCGCCCGUAUCUCGGUAUUCCGUAUCUUAAUCG ...(((((((.....)))).)))...---......------.....(((((((((((((((((((((.(...............).))))))))))).....)).)))))..))) ( -23.36) >consensus AUUGUGUAACUAACCGUUGUCACCGUUGUCAUUAAA_CAUAAAAAACGAAAGAUCGUCGAGAUGCGGUCUUUAAACUUAAUUACGCCCGUAUCUCGGUAUUCCGUAUCUUAAUCG ...(((((((.....)))).))).......................(((((((((((((((((((((.(...............).))))))))))).....)).)))))..))) (-20.98 = -21.82 + 0.84)

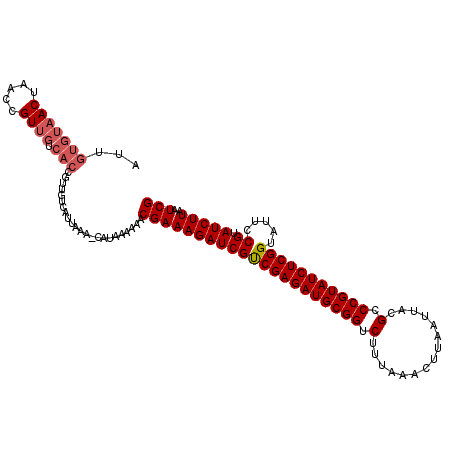
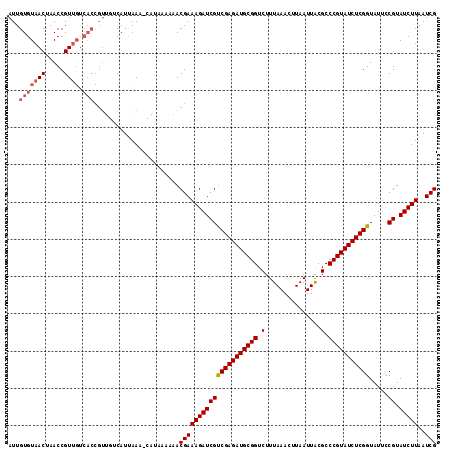
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:47 2006