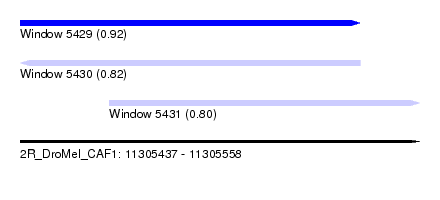
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,305,437 – 11,305,558 |
| Length | 121 |
| Max. P | 0.923000 |

| Location | 11,305,437 – 11,305,540 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 87.18 |
| Mean single sequence MFE | -35.94 |
| Consensus MFE | -27.44 |
| Energy contribution | -27.56 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11305437 103 + 20766785 UGCGGGAAAUAACCAAGACGUGUGGGCCACACGAUGGUAGUGCUAAGCU-AACAGUGGAUUGGGAAAUUUUCGGGAACGGGAUACUUUCCCAACCUGCUG---CUUC .(((((......(((...((((((...)))))).((.((((.....)))-).)).))).((((((((.(..((....))..)...)))))))))))))..---.... ( -35.80) >DroSec_CAF1 92668 106 + 1 UGCGGGAAAUAACCAAGACGUGUGGGCCACACGAUGGUAGAGCCAAGCU-AACAGUGGAUUGGGAAAUUUUCGGGAACGAGAUACUUUCCCAACCUGCUGCUGCUGC .((.((.....((((...((((((...)))))).))))....)).(((.-..(((..(.((((((((.(((((....)))))...)))))))).)..)))..))))) ( -39.80) >DroSim_CAF1 96349 106 + 1 UGCGGGAAAUAACCAAGACGUGUGGGCCACACGAUGGUAGUGCCAAGCU-AACAGUGGAUUGGGAAAUUUUCGGGAACGAGAUACUUUCCCAACCUGCUGCUGCUGC .((((.(....((((...((((((...)))))).))))..).)).(((.-..(((..(.((((((((.(((((....)))))...)))))))).)..)))..))))) ( -40.30) >DroEre_CAF1 93890 95 + 1 UGCGGGAAAUAACCAAGACGUGUGGGCCACACUCUGUCAGAG-----CU-ACCAGUGGAUUGGGAAAAUUUCCGGAACGGGAUACUUUCCCAACCUGCUG------U .((((.......))..((((.(((.....)))..))))...)-----).-..(((..(.((((((((..(((((...)))))...)))))))).)..)))------. ( -32.20) >DroYak_CAF1 95012 100 + 1 UGCGGGAAAUAACCAAGACGUGUGGGCCACACUCUUGCAGAG-----CUCACCAGUGGAUUGGGAAAUUUUCGGUAACAGGAUACUUUCCCAACCUGCUGCU--UGC .(((((.......(((((.(((((...)))))))))).....-----.....(((..(.((((((((..(((.......)))...)))))))).)..)))))--))) ( -31.60) >consensus UGCGGGAAAUAACCAAGACGUGUGGGCCACACGAUGGUAGAGC_AAGCU_AACAGUGGAUUGGGAAAUUUUCGGGAACGGGAUACUUUCCCAACCUGCUGCU_CUGC (((((.......)).....(((((...)))))....))).............(((..(.((((((((.(((((....)))))...)))))))).)..)))....... (-27.44 = -27.56 + 0.12)

| Location | 11,305,437 – 11,305,540 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.18 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -23.19 |
| Energy contribution | -24.15 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11305437 103 - 20766785 GAAG---CAGCAGGUUGGGAAAGUAUCCCGUUCCCGAAAAUUUCCCAAUCCACUGUU-AGCUUAGCACUACCAUCGUGUGGCCCACACGUCUUGGUUAUUUCCCGCA ..((---(((..((((((((((...((........))...))))))))))..)))))-.((........((((.((((((...))))))...))))........)). ( -32.09) >DroSec_CAF1 92668 106 - 1 GCAGCAGCAGCAGGUUGGGAAAGUAUCUCGUUCCCGAAAAUUUCCCAAUCCACUGUU-AGCUUGGCUCUACCAUCGUGUGGCCCACACGUCUUGGUUAUUUCCCGCA ((((((((((..((((((((((.....(((....)))...))))))))))..)))))-.))).((....((((.((((((...))))))...))))......)))). ( -38.20) >DroSim_CAF1 96349 106 - 1 GCAGCAGCAGCAGGUUGGGAAAGUAUCUCGUUCCCGAAAAUUUCCCAAUCCACUGUU-AGCUUGGCACUACCAUCGUGUGGCCCACACGUCUUGGUUAUUUCCCGCA ((((((((((..((((((((((.....(((....)))...))))))))))..)))))-.))).((....((((.((((((...))))))...))))......)))). ( -38.20) >DroEre_CAF1 93890 95 - 1 A------CAGCAGGUUGGGAAAGUAUCCCGUUCCGGAAAUUUUCCCAAUCCACUGGU-AG-----CUCUGACAGAGUGUGGCCCACACGUCUUGGUUAUUUCCCGCA .------(((..((((((((((((.(((......))).))))))))))))..)))((-((-----((..(((...(((((...))))))))..))))))........ ( -34.50) >DroYak_CAF1 95012 100 - 1 GCA--AGCAGCAGGUUGGGAAAGUAUCCUGUUACCGAAAAUUUCCCAAUCCACUGGUGAG-----CUCUGCAAGAGUGUGGCCCACACGUCUUGGUUAUUUCCCGCA ((.--..(((..((((((((((...((........))...))))))))))..))).....-----.....((((((((((...))))).)))))..........)). ( -29.50) >consensus GCAG_AGCAGCAGGUUGGGAAAGUAUCCCGUUCCCGAAAAUUUCCCAAUCCACUGUU_AGCUU_GCUCUACCAUCGUGUGGCCCACACGUCUUGGUUAUUUCCCGCA .......(((..((((((((((...((........))...))))))))))..)))....((........((((.((((((...))))))...))))........)). (-23.19 = -24.15 + 0.96)

| Location | 11,305,464 – 11,305,558 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 84.70 |
| Mean single sequence MFE | -33.84 |
| Consensus MFE | -22.57 |
| Energy contribution | -22.89 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.797080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11305464 94 + 20766785 CACACGAUGGUAGUGCUAAGCU-AACAGUGGAUUGGGAAAUUUUCGGGAACGGGAUACUUUCCCAACCUGCUG---CUUCUACCUGCUGGGCGGCGCA ............(((((..((.-..(((..(.((((((((.(..((....))..)...)))))))).)..)))---......((....))))))))). ( -32.50) >DroSec_CAF1 92695 97 + 1 CACACGAUGGUAGAGCCAAGCU-AACAGUGGAUUGGGAAAUUUUCGGGAACGAGAUACUUUCCCAACCUGCUGCUGCUGCUACCUGCUGGGCGGCGCA .......(((.....))).((.-..(((..(.((((((((.(((((....)))))...)))))))).)..)))..((((((........)))))))). ( -37.70) >DroSim_CAF1 96376 97 + 1 CACACGAUGGUAGUGCCAAGCU-AACAGUGGAUUGGGAAAUUUUCGGGAACGAGAUACUUUCCCAACCUGCUGCUGCUGCUACCUGCUGGGCGGCGCA ............(((((.(((.-..(((..(.((((((((.(((((....)))))...)))))))).)..)))..)))(((........)))))))). ( -39.40) >DroEre_CAF1 93917 84 + 1 CACACUCUGUCAGAG-----CU-ACCAGUGGAUUGGGAAAAUUUCCGGAACGGGAUACUUUCCCAACCUGCUG------UUACCUGCUGGGCGG--CA ......(((((..((-----(.-..(((..(.((((((((..(((((...)))))...)))))))).)..)))------......))).)))))--.. ( -29.80) >DroYak_CAF1 95039 89 + 1 CACACUCUUGCAGAG-----CUCACCAGUGGAUUGGGAAAUUUUCGGUAACAGGAUACUUUCCCAACCUGCUGCU--UGCUACCUGCUGGGCGG--CA ..(.(((..((((((-----(....(((..(.((((((((..(((.......)))...)))))))).)..)))..--.)))..)))).))).).--.. ( -29.80) >consensus CACACGAUGGUAGAGC_AAGCU_AACAGUGGAUUGGGAAAUUUUCGGGAACGGGAUACUUUCCCAACCUGCUGCU_CUGCUACCUGCUGGGCGGCGCA .......((((((.(..........(((..(.((((((((.(((((....)))))...)))))))).)..))).........)))))))......... (-22.57 = -22.89 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:36 2006