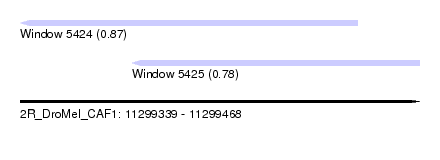
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,299,339 – 11,299,468 |
| Length | 129 |
| Max. P | 0.874052 |

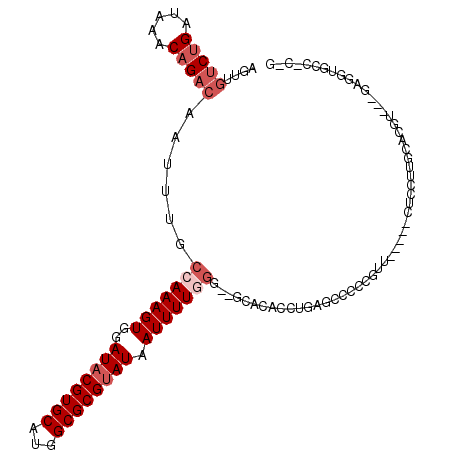
| Location | 11,299,339 – 11,299,448 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.81 |
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -17.20 |
| Energy contribution | -18.37 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11299339 109 - 20766785 AGUUGUCUGAUAAACAGACAAUUUGCCAAAGUGGAUACGUGCAUGGCGCGUAUAAUUUUGGG--GCACACCUGAGCCCCCGUUCGGACCUCCUUUCGCAU---CAGGUCCC-CCC (((((((((.....)))))))))....(((((..((((((((...)))))))).)))))(((--((........))))).....((((((..........---.)))))).-... ( -37.80) >DroVir_CAF1 80895 103 - 1 GGUAGUCGGAUAAACAGACAAUUUGCCAAAGUGGAUACGUGCAGGGCGCGCAUAAUUUUCGGCCGUACACCUGAGCCUAGC-------CACGUGGCAGG-C--UCAGAGUG-G-G ....(((.........))).....((((((((..((.(((((...))))).)).))))).)))....(((((((((((.((-------(....))))))-)--)))).)))-.-. ( -38.30) >DroGri_CAF1 65056 92 - 1 GAAAGUCUGAUAAACAGACAAUUUGGCAAAGUGGAUACGUGCAUGGCGCGUAUAAUUUUGGGCCGCACACCUGAGCCAAGG-------CACGUGGCAAG---------------- ....(((((.....))))).......((((((..((((((((...)))))))).)))))).(((((...(((......)))-------...)))))...---------------- ( -32.00) >DroWil_CAF1 143928 99 - 1 ------CUGAUAAACAGACAAUUUGCCAAAGUGGAUACGUGCAUGGCGCGUAUAAUUUUAGG--CCACACCUGAGCCCCGAGU-----CUCCUAGUCUGU---GAGUUUCCGUGU ------..(((..((((((.....((((((((..((((((((...)))))))).))))).))--).......((((.....).-----)))...))))))---..)))....... ( -25.70) >DroYak_CAF1 88541 112 - 1 AGUUGUCUGAUAAACAGACAAUUUGCCAAAGUGGAUACGUGCAUGGCGCGUAUAAUUUUGGG--GCACACCUGAGCCCCCGUUCGGACCUCCUUUCGCACAUCGGAGUACC-CCU (((((((((.....)))))))))....(((((..((((((((...)))))))).)))))(((--(.....((((((....))))))..((((...........))))..))-)). ( -36.50) >DroAna_CAF1 80157 104 - 1 AGUUGUCUGAUAAACAGACAAUUUGCCAAAGUGGAUACGUGCAUGGCGCGUAUAAUUUUGGG--GCGCACCUGAGCCCGCUUU-----CUCCUUUCCCCUU--UCGGUGCC-C-G (((((((((.....)))))))))..(((((((..((((((((...)))))))).)))))))(--(.(((((.(((........-----............)--))))))))-)-. ( -36.35) >consensus AGUUGUCUGAUAAACAGACAAUUUGCCAAAGUGGAUACGUGCAUGGCGCGUAUAAUUUUGGG__GCACACCUGAGCCCCCGUU_____CUCCUUGCACGU___GAGGUGCC_C_G ....(((((.....)))))......(((((((..((((((((...)))))))).)))))))...................................................... (-17.20 = -18.37 + 1.17)

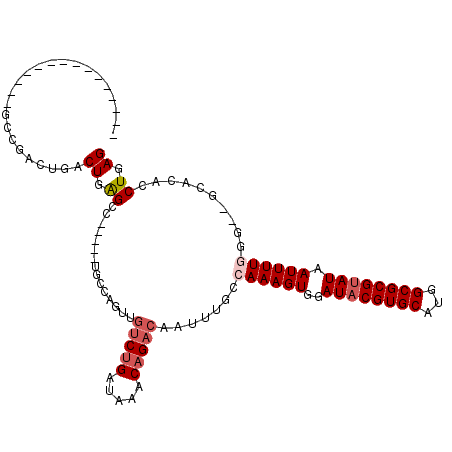
| Location | 11,299,375 – 11,299,468 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.16 |
| Mean single sequence MFE | -32.56 |
| Consensus MFE | -16.38 |
| Energy contribution | -17.13 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11299375 93 - 20766785 ---------------ACUGACUGACUGGGCC-----UGCCAGUUGUCUGAUAAACAGACAAUUUGCCAAAGUGGAUACGUGCAUGGCGCGUAUAAUUUUGGG--GCACACCUGAG ---------------...........(((..-----(((((((((((((.....)))))))))..(((((((..((((((((...)))))))).))))))))--)))..)))... ( -36.70) >DroVir_CAF1 80923 100 - 1 ---------------GUCGACUGACUGGGCUGCCGGGGCUGGUAGUCGGAUAAACAGACAAUUUGCCAAAGUGGAUACGUGCAGGGCGCGCAUAAUUUUCGGCCGUACACCUGAG ---------------.(((((((.(..(.((....)).)..)))))))).....(((.......((((((((..((.(((((...))))).)).))))).))).......))).. ( -27.74) >DroPse_CAF1 93181 107 - 1 CCGACAGGCAGACGGUCUGACUGACUGGGCC-----UGCCAGUUGUCUGAUAAACAGACAAUUUGCCAAAGUGGAUACGUGCAUGGCGCGUAUAAUUUUG-G--GCACACCUGAG ..(.(((((...(((((.....))))).)))-----)).)(((((((((.....)))))))))(((((((((..((((((((...)))))))).))))).-)--)))........ ( -43.50) >DroGri_CAF1 65073 91 - 1 ---------------GGCGACUGACUGAG---------CUGAAAGUCUGAUAAACAGACAAUUUGGCAAAGUGGAUACGUGCAUGGCGCGUAUAAUUUUGGGCCGCACACCUGAG ---------------(((..((.(((..(---------(..((.(((((.....)))))..))..))..)))))((((((((...))))))))........)))........... ( -27.80) >DroWil_CAF1 143960 78 - 1 ---------------CUC----GACUGAGC----------------CUGAUAAACAGACAAUUUGCCAAAGUGGAUACGUGCAUGGCGCGUAUAAUUUUAGG--CCACACCUGAG ---------------(((----(..((.((----------------((((...............((.....))((((((((...))))))))....)))))--)..))..)))) ( -20.40) >DroPer_CAF1 92464 107 - 1 CCGACAGACAGACGGUAUGACUGACUGAGCC-----UGCCAGUUGUCUGAUAAACAGACAAUUUGCCAAAGUGGAUACGUGCAUGGCGCGUAUAAUUUUG-G--GCACACCUGAG ....(((.(((.(((.....))).))).(((-----.((.(((((((((.....))))))))).))((((((..((((((((...)))))))).))))))-)--))....))).. ( -39.20) >consensus _______________GCCGACUGACUGAGCC_____UGCCAGUUGUCUGAUAAACAGACAAUUUGCCAAAGUGGAUACGUGCAUGGCGCGUAUAAUUUUGGG__GCACACCUGAG ........................((.((...............(((((.....))))).......((((((..((((((((...)))))))).))))))..........)).)) (-16.38 = -17.13 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:31 2006