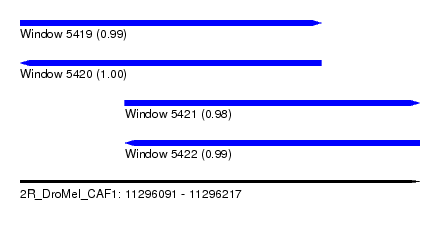
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,296,091 – 11,296,217 |
| Length | 126 |
| Max. P | 0.999936 |

| Location | 11,296,091 – 11,296,186 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -24.70 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11296091 95 + 20766785 AAAACACGAAUCGCAGGAGUAAAUGAAACCAAGAGCCCUUGAGUUCAGCUUGUAGCAUACUUUCAGGCAUGAACACAGCGCAGCCCGAAAGUAUG ............(((((......((((..((((....))))..)))).)))))..(((((((((.(((.((.........))))).))))))))) ( -25.20) >DroSec_CAF1 83313 95 + 1 AAAACACGAAACGCAGGAGUGAAUGAAACCAAGAGCCCUUGAGCUCAGCGUGUAGCAUACUUUCAGGCAUGCACACAGCGCAGCCCGAAAGUAUG ...(((((...(((....)))...........((((......))))..)))))..(((((((((.(((.(((.......)))))).))))))))) ( -31.80) >DroSim_CAF1 86970 95 + 1 AAAACACGAAACGCAGGAGUAAAUGAAACCAAGAGCCCUUGAGCUCAGCGUGUAGCAUACUUUCAGGCAUGCACACAGCGCAGCCCGAAAGUAUG ...(((((.......((...........))..((((......))))..)))))..(((((((((.(((.(((.......)))))).))))))))) ( -30.10) >DroEre_CAF1 84443 95 + 1 AAAACUGGAAACGCAGGAGUAAAUGAAACCAAGAGCUCUUGUGCUCGGCUUGUAGCAUACUUUCAGGCGUGCACACAGCGCAGCCCGAAAGUAUG ....((((...(((((((((...((....))...))))))))).)))).......(((((((((.(((((((.....)))).))).))))))))) ( -34.10) >DroYak_CAF1 85295 95 + 1 AAAACCCGAAACGCAGGAGUAAAUGAAACCAAGAGCUCUUGAGCUCAGCGCGUAGCAUACUUUCAGGCGUGCACACAGCACAGCCCGAAAGUAUG ..........((((.((...........))..(((((....)))))...))))..(((((((((.(((((((.....)))).))).))))))))) ( -34.80) >consensus AAAACACGAAACGCAGGAGUAAAUGAAACCAAGAGCCCUUGAGCUCAGCGUGUAGCAUACUUUCAGGCAUGCACACAGCGCAGCCCGAAAGUAUG ...(((((.......((...........))..((((......))))..)))))..(((((((((.(((.(((.......)))))).))))))))) (-24.70 = -25.30 + 0.60)

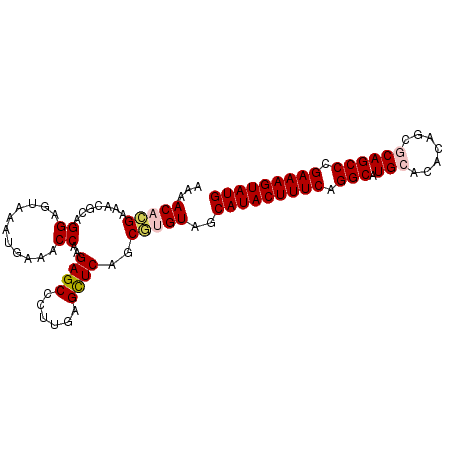
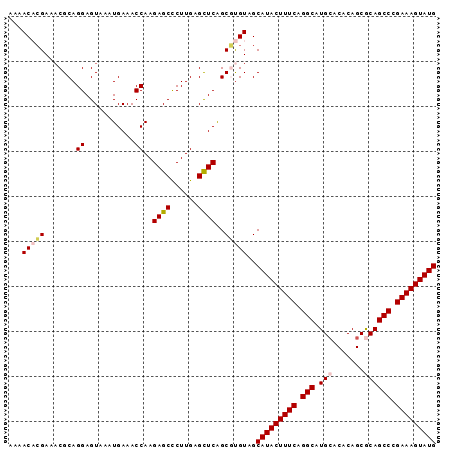
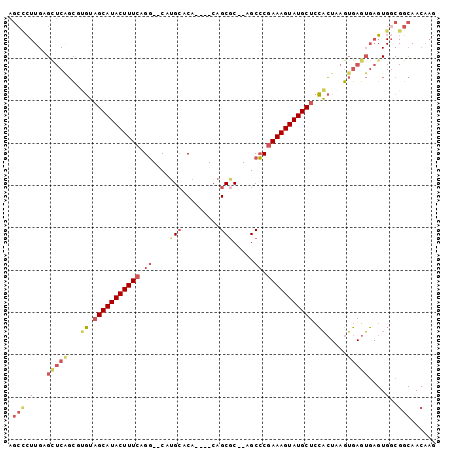
| Location | 11,296,091 – 11,296,186 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -30.64 |
| Energy contribution | -31.48 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.67 |
| SVM RNA-class probability | 0.999936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11296091 95 - 20766785 CAUACUUUCGGGCUGCGCUGUGUUCAUGCCUGAAAGUAUGCUACAAGCUGAACUCAAGGGCUCUUGGUUUCAUUUACUCCUGCGAUUCGUGUUUU (((((((((((((((.((...)).)).)))))))))))))....((((((((..((((....))))..)))).........(((...))))))). ( -26.00) >DroSec_CAF1 83313 95 - 1 CAUACUUUCGGGCUGCGCUGUGUGCAUGCCUGAAAGUAUGCUACACGCUGAGCUCAAGGGCUCUUGGUUUCAUUCACUCCUGCGUUUCGUGUUUU ((((((((((((((((((...))))).)))))))))))))..(((((..(((((....))))).(((......)))...........)))))... ( -37.00) >DroSim_CAF1 86970 95 - 1 CAUACUUUCGGGCUGCGCUGUGUGCAUGCCUGAAAGUAUGCUACACGCUGAGCUCAAGGGCUCUUGGUUUCAUUUACUCCUGCGUUUCGUGUUUU ((((((((((((((((((...))))).)))))))))))))..(((((..(((((....)))))..((...........)).......)))))... ( -36.90) >DroEre_CAF1 84443 95 - 1 CAUACUUUCGGGCUGCGCUGUGUGCACGCCUGAAAGUAUGCUACAAGCCGAGCACAAGAGCUCUUGGUUUCAUUUACUCCUGCGUUUCCAGUUUU ((((((((((((((((((...))))).)))))))))))))....(((((((((......))))..))))).........(((......))).... ( -34.40) >DroYak_CAF1 85295 95 - 1 CAUACUUUCGGGCUGUGCUGUGUGCACGCCUGAAAGUAUGCUACGCGCUGAGCUCAAGAGCUCUUGGUUUCAUUUACUCCUGCGUUUCGGGUUUU (((((((((((((.((((.....)))))))))))))))))..((((...(((((....)))))..((...........)).)))).......... ( -37.40) >consensus CAUACUUUCGGGCUGCGCUGUGUGCAUGCCUGAAAGUAUGCUACACGCUGAGCUCAAGGGCUCUUGGUUUCAUUUACUCCUGCGUUUCGUGUUUU ((((((((((((((((((...))))).)))))))))))))....((((.(((((....)))))..((...........)).)))).......... (-30.64 = -31.48 + 0.84)

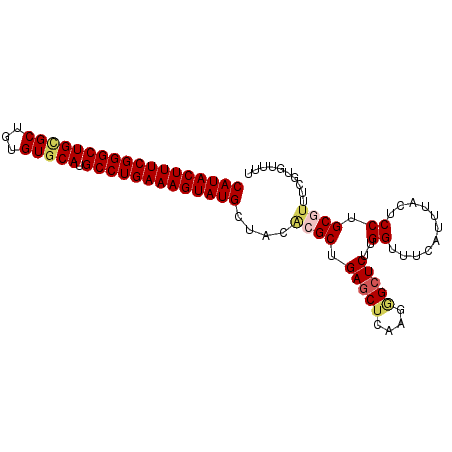
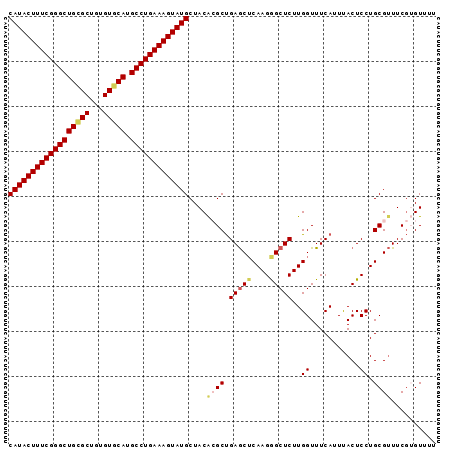
| Location | 11,296,124 – 11,296,217 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -18.79 |
| Energy contribution | -20.67 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11296124 93 + 20766785 AGCCCUUGAGUUCAGCUUGUAGCAUACUUUCAGG--CAUGAACA----CAGCGC--AGCCCGAAAGUAUGCUCCACUAAGUGAGUGAGUGGCGGCAACAAG .(((((((...(((.((((((((((((((((.((--(.((....----.....)--)))).)))))))))))..)).)))))).)))).)))(....)... ( -32.20) >DroVir_CAF1 77545 86 + 1 ACACU--------UUUCCAUUGCAUACUUUCGGCUGCAAGCGCACAAAGCGCGCGGCGCGCAAAAGUAUGCUGUGAUAAAUGAGUGCG-----GC--CAAC .((((--------(...(((.((((((((((((((((..((((.....))))))))).))..))))))))).)))......)))))..-----..--.... ( -32.30) >DroSec_CAF1 83346 93 + 1 AGCCCUUGAGCUCAGCGUGUAGCAUACUUUCAGG--CAUGCACA----CAGCGC--AGCCCGAAAGUAUGCUCCACUAAGUGAGUGAGUGCCGGCAACAAG .(((.....(((((..(((.(((((((((((.((--(.(((...----....))--)))).))))))))))).)))....))))).......)))...... ( -38.10) >DroSim_CAF1 87003 93 + 1 AGCCCUUGAGCUCAGCGUGUAGCAUACUUUCAGG--CAUGCACA----CAGCGC--AGCCCGAAAGUAUGCUCCACUAAGUGAGUGAGUGGCGGCAACAAG .((((((..(((((..(((.(((((((((((.((--(.(((...----....))--)))).))))))))))).)))....)))))))).)))(....)... ( -41.80) >DroEre_CAF1 84476 93 + 1 AGCUCUUGUGCUCGGCUUGUAGCAUACUUUCAGG--CGUGCACA----CAGCGC--AGCCCGAAAGUAUGCUCCACUAAGUGAGUGAGUGGCGGCAACAAG .((((((..(((((.((((((((((((((((.((--(((((...----..))))--.))).)))))))))))..)).))))))))))).)))(....)... ( -39.40) >DroYak_CAF1 85328 93 + 1 AGCUCUUGAGCUCAGCGCGUAGCAUACUUUCAGG--CGUGCACA----CAGCAC--AGCCCGAAAGUAUGCUCCACUAAGUGAGUGAGUGGCGGCAACAAG ((((....))))..((.((((((((((((((.((--(((((...----..))))--.))).))))))))))).((((.....))))....)))))...... ( -38.10) >consensus AGCCCUUGAGCUCAGCGUGUAGCAUACUUUCAGG__CAUGCACA____CAGCGC__AGCCCGAAAGUAUGCUCCACUAAGUGAGUGAGUGGCGGCAACAAG .(((((...(((((...((.(((((((((((.((....(((.........))).....)).))))))))))).)).....)))))....)).)))...... (-18.79 = -20.67 + 1.88)

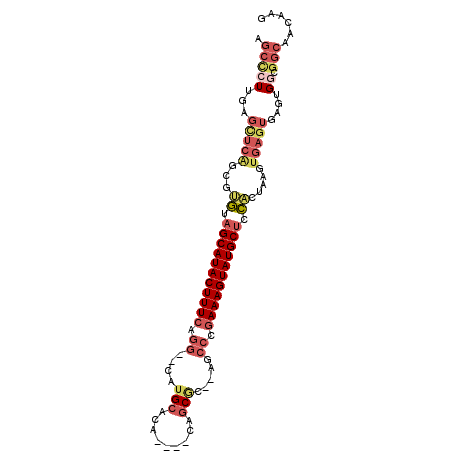
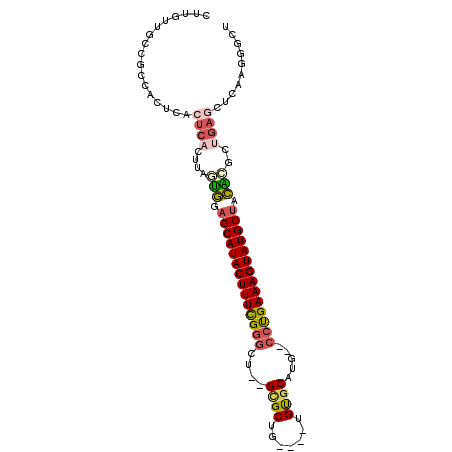
| Location | 11,296,124 – 11,296,217 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -41.13 |
| Consensus MFE | -26.63 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.32 |
| Mean z-score | -4.49 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11296124 93 - 20766785 CUUGUUGCCGCCACUCACUCACUUAGUGGAGCAUACUUUCGGGCU--GCGCUG----UGUUCAUG--CCUGAAAGUAUGCUACAAGCUGAACUCAAGGGCU ......(((.(((((.........)))))((((((((((((((((--(.((..----.)).)).)--))))))))))))))................))). ( -35.00) >DroVir_CAF1 77545 86 - 1 GUUG--GC-----CGCACUCAUUUAUCACAGCAUACUUUUGCGCGCCGCGCGCUUUGUGCGCUUGCAGCCGAAAGUAUGCAAUGGAAA--------AGUGU ....--..-----.(((((..(((((....(((((((((((.((((.((((((...))))))..)).))))))))))))).)))))..--------))))) ( -33.10) >DroSec_CAF1 83346 93 - 1 CUUGUUGCCGGCACUCACUCACUUAGUGGAGCAUACUUUCGGGCU--GCGCUG----UGUGCAUG--CCUGAAAGUAUGCUACACGCUGAGCUCAAGGGCU ((((..((((((..(((((.....)))))((((((((((((((((--((((..----.))))).)--))))))))))))))....)))).)).)))).... ( -46.40) >DroSim_CAF1 87003 93 - 1 CUUGUUGCCGCCACUCACUCACUUAGUGGAGCAUACUUUCGGGCU--GCGCUG----UGUGCAUG--CCUGAAAGUAUGCUACACGCUGAGCUCAAGGGCU .........(((.((......(((((((.((((((((((((((((--((((..----.))))).)--))))))))))))))...)))))))....))))). ( -45.50) >DroEre_CAF1 84476 93 - 1 CUUGUUGCCGCCACUCACUCACUUAGUGGAGCAUACUUUCGGGCU--GCGCUG----UGUGCACG--CCUGAAAGUAUGCUACAAGCCGAGCACAAGAGCU (((((.(((((((((.........)))))((((((((((((((((--((((..----.))))).)--))))))))))))))......)).))))))).... ( -44.00) >DroYak_CAF1 85328 93 - 1 CUUGUUGCCGCCACUCACUCACUUAGUGGAGCAUACUUUCGGGCU--GUGCUG----UGUGCACG--CCUGAAAGUAUGCUACGCGCUGAGCUCAAGAGCU .............(((.....(((((((((((((((((((((((.--((((..----...)))))--)))))))))))))).).))))))).....))).. ( -42.80) >consensus CUUGUUGCCGCCACUCACUCACUUAGUGGAGCAUACUUUCGGGCU__GCGCUG____UGUGCAUG__CCUGAAAGUAUGCUACACGCUGAGCUCAAGGGCU .................((((....(((.((((((((((((((....((((.......)))).....)))))))))))))).)))..)))).......... (-26.63 = -27.30 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:28 2006