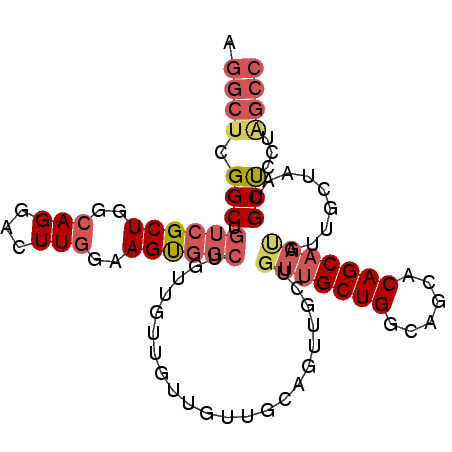
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,250,786 – 11,250,882 |
| Length | 96 |
| Max. P | 0.584658 |

| Location | 11,250,786 – 11,250,882 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -15.68 |
| Energy contribution | -16.82 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11250786 96 + 20766785 GGCUAGAAGCAUUAGCAACAUUGCUGUGCUGCCAGCAACAGCAACUGCAACAACAACAACAGCCACUUCCAAGUCCUGCCAGCGACAGCCGAGCCU ((((.(..((....(((...(((((((((.....)).))))))).)))...........(((..(((....))).)))...))..)))))...... ( -24.40) >DroSec_CAF1 37490 96 + 1 GGCUAGGAGCAUCAGCAACAUUGCUGUGCUGCCAGCAACAGCAACUGCAACAACAACAACAGCCACUUCCAAGUCCUGCCAGCGACAGCCGAGCCU ((((.((((((.(((((....))))))))).)).(((........)))............)))).............(((.((....)).).)).. ( -26.20) >DroSim_CAF1 40163 96 + 1 GGCUAGGAGCAUCAGCAACAUUGCUGUGCUGCCAGCAACAGCAACUGCAACAACAACAACAGCCACUUCCAAGUCCUGCCAGCGACAGCCGAGCCU ((((.((((((.(((((....))))))))).)).(((........)))............)))).............(((.((....)).).)).. ( -26.20) >DroYak_CAF1 38868 96 + 1 GGCUAGGAGCAUCAGCAACAUUGCUGUGCUGCCAGCAACAGCAACAACCACAACAGCAACAGCCACUUCCAAGUCCUGCCAGCGACAGCCGAGCCU ((((.((((((.(((((....))))))))).)).((....))..................)))).............(((.((....)).).)).. ( -25.10) >DroAna_CAF1 35668 84 + 1 GGCCAAGAGCGUUACCCAUGUUGCUGUGUUGCCAGCAACGCCAACGGCCA------------CCGCUAACAAGUCCUGCCAGCGACAGCCGAGCCU (((................(((((((......))))))).....((((..------------.((((..((.....))..))))...)))).))). ( -26.10) >DroPer_CAF1 40176 78 + 1 ------GGGCGUUAGCAACAUUGCUGUGCUGGCAGCGU------------CAACGGCGACUGGGACUGCAAAGUCCUGCCAGCGUCAGCCGAGUCU ------.(((..(((((....))))).(((((((((((------------.....))).))((((((....))))))))))))....)))...... ( -33.90) >consensus GGCUAGGAGCAUCAGCAACAUUGCUGUGCUGCCAGCAACAGCAACUGCAACAACAACAACAGCCACUUCCAAGUCCUGCCAGCGACAGCCGAGCCU ((((....((....))......(((((((((.(((.....((....))................(((....))).))).))))).))))..)))). (-15.68 = -16.82 + 1.14)

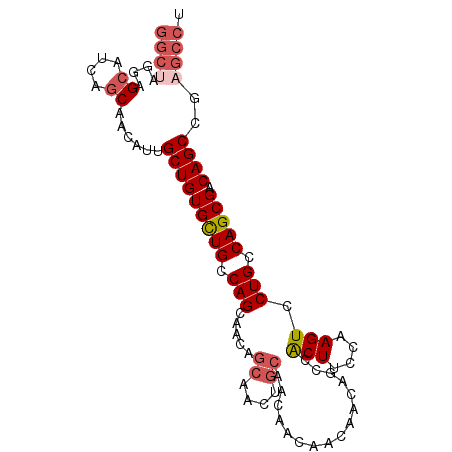
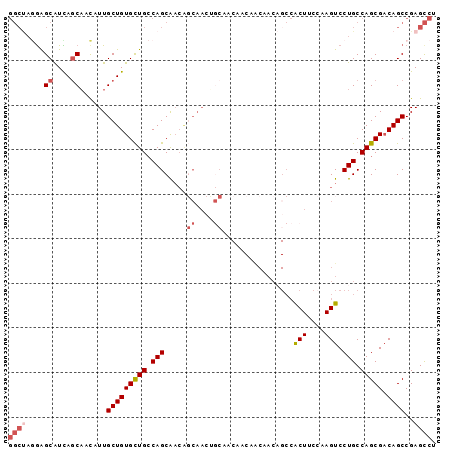
| Location | 11,250,786 – 11,250,882 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -36.07 |
| Consensus MFE | -18.71 |
| Energy contribution | -20.05 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11250786 96 - 20766785 AGGCUCGGCUGUCGCUGGCAGGACUUGGAAGUGGCUGUUGUUGUUGUUGCAGUUGCUGUUGCUGGCAGCACAGCAAUGUUGCUAAUGCUUCUAGCC ..((.((((....)))))).((.((.(((((((((.((((((((.(((((....((....))..)))))))))))))...)))...)))))))))) ( -35.60) >DroSec_CAF1 37490 96 - 1 AGGCUCGGCUGUCGCUGGCAGGACUUGGAAGUGGCUGUUGUUGUUGUUGCAGUUGCUGUUGCUGGCAGCACAGCAAUGUUGCUGAUGCUCCUAGCC .((((.((.....((..((((........((..(((((..........)))))..)).))))..))(((((((((....))))).)))))).)))) ( -36.90) >DroSim_CAF1 40163 96 - 1 AGGCUCGGCUGUCGCUGGCAGGACUUGGAAGUGGCUGUUGUUGUUGUUGCAGUUGCUGUUGCUGGCAGCACAGCAAUGUUGCUGAUGCUCCUAGCC .((((.((.....((..((((........((..(((((..........)))))..)).))))..))(((((((((....))))).)))))).)))) ( -36.90) >DroYak_CAF1 38868 96 - 1 AGGCUCGGCUGUCGCUGGCAGGACUUGGAAGUGGCUGUUGCUGUUGUGGUUGUUGCUGUUGCUGGCAGCACAGCAAUGUUGCUGAUGCUCCUAGCC .((((.((.....((..((((........((..((..(..(....)..)..))..)).))))..))(((((((((....))))).)))))).)))) ( -35.60) >DroAna_CAF1 35668 84 - 1 AGGCUCGGCUGUCGCUGGCAGGACUUGUUAGCGG------------UGGCCGUUGGCGUUGCUGGCAACACAGCAACAUGGGUAACGCUCUUGGCC .((((((((..((((((((((...))))))))))------------..))))..((((((((((....).((......)))))))))))...)))) ( -40.00) >DroPer_CAF1 40176 78 - 1 AGACUCGGCUGACGCUGGCAGGACUUUGCAGUCCCAGUCGCCGUUG------------ACGCUGCCAGCACAGCAAUGUUGCUAACGCCC------ .(((...((((..((((((((((((....)))))..((((....))------------))..))))))).))))...)))((....))..------ ( -31.40) >consensus AGGCUCGGCUGUCGCUGGCAGGACUUGGAAGUGGCUGUUGUUGUUGUUGCAGUUGCUGUUGCUGGCAGCACAGCAAUGUUGCUAAUGCUCCUAGCC .((((.(((.((((((..(((...)))..))))))......................(((((((......))))))).........)))...)))) (-18.71 = -20.05 + 1.34)

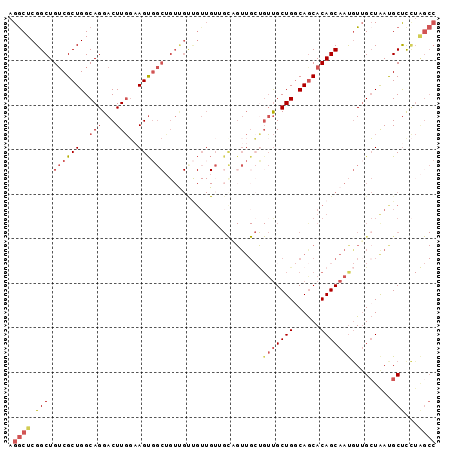
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:13 2006