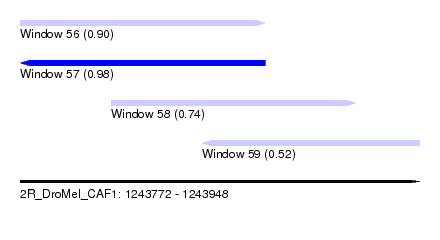
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,243,772 – 1,243,948 |
| Length | 176 |
| Max. P | 0.975231 |

| Location | 1,243,772 – 1,243,880 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -39.34 |
| Consensus MFE | -26.76 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1243772 108 + 20766785 GGAGGCAAACAUAGUCCUGGAAGGGCGAGCACUUGAGAUAAUCGCCUUAGGGUUGUUUUGGCAGCGGGUGCUACCAGGAGACUGCUACCAGGC------------GGUUGCCAUAAACGA ...(((........((((((.((.((..((..(..((((((((.......))))))))..)..))..)).)).))))))(((((((....)))------------)))))))........ ( -39.60) >DroSim_CAF1 1 104 + 1 ----------------CUGGAAGGGCGAGCACUUGAGAUAAUCGCCUUAGGGUUGUUUUGGCAGCGGGUGCUACCAGGAGACUGCUACCAGGCCCGGGUUGCCCCGCUCGCCAUAAACGA ----------------.....(((((((.............)))))))....((((((((((((((((.((.(((.((.(.(((....))).))).))).)))))))).)))).)))))) ( -42.72) >DroYak_CAF1 1 97 + 1 --------------------AAGGGCGAGCACUUGAGAUAAUCGCCUUAGGGUUGUUUUGGUAGCGGGUGCUACCAGGAGACUACUACCAGGC---GGUUGCCCCGUUCACCAUAAACGA --------------------..((((((.(....((.....))(((((((((((.((((((((((....)))))))))))))).)))..))))---).))))))((((.......)))). ( -35.70) >consensus ________________CUGGAAGGGCGAGCACUUGAGAUAAUCGCCUUAGGGUUGUUUUGGCAGCGGGUGCUACCAGGAGACUGCUACCAGGC___GGUUGCCCCGCUCGCCAUAAACGA .......................(((((((....((.....))(((((((((((.((((((.(((....))).)))))))))).)))..))))............)))))))........ (-26.76 = -27.10 + 0.34)

| Location | 1,243,772 – 1,243,880 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -25.26 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1243772 108 - 20766785 UCGUUUAUGGCAACC------------GCCUGGUAGCAGUCUCCUGGUAGCACCCGCUGCCAAAACAACCCUAAGGCGAUUAUCUCAAGUGCUCGCCCUUCCAGGACUAUGUUUGCCUCC ........(((((.(------------((((((.((........(((((((....)))))))............(((((.(((.....))).))))))).)))))....)).)))))... ( -33.50) >DroSim_CAF1 1 104 - 1 UCGUUUAUGGCGAGCGGGGCAACCCGGGCCUGGUAGCAGUCUCCUGGUAGCACCCGCUGCCAAAACAACCCUAAGGCGAUUAUCUCAAGUGCUCGCCCUUCCAG---------------- ..((((.((((.((((((((.(((.(((.(((....))).).)).))).)).))))))))))))))........(((((.(((.....))).))))).......---------------- ( -41.80) >DroYak_CAF1 1 97 - 1 UCGUUUAUGGUGAACGGGGCAACC---GCCUGGUAGUAGUCUCCUGGUAGCACCCGCUACCAAAACAACCCUAAGGCGAUUAUCUCAAGUGCUCGCCCUU-------------------- ..((((..((.((((.((((....---)))).)).....)).))(((((((....)))))))))))........(((((.(((.....))).)))))...-------------------- ( -30.50) >consensus UCGUUUAUGGCGAACGGGGCAACC___GCCUGGUAGCAGUCUCCUGGUAGCACCCGCUGCCAAAACAACCCUAAGGCGAUUAUCUCAAGUGCUCGCCCUUCCAG________________ ..((((..(((....(((....)))..))).((.((....))))(((((((....)))))))))))........(((((.(((.....))).)))))....................... (-25.26 = -25.40 + 0.14)

| Location | 1,243,812 – 1,243,920 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -47.37 |
| Consensus MFE | -37.59 |
| Energy contribution | -40.37 |
| Covariance contribution | 2.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
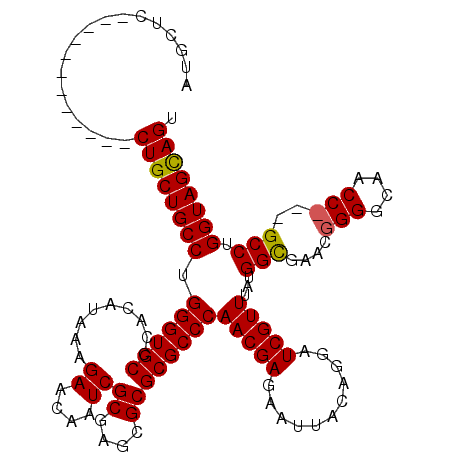
>2R_DroMel_CAF1 1243812 108 + 20766785 AUCGCCUUAGGGUUGUUUUGGCAGCGGGUGCUACCAGGAGACUGCUACCAGGC------------GGUUGCCAUAAACGAUCCUGUAAUUCUCGUUGGGCGCGCGCUCGCGAUUGUUCUU (((((..((((((((((((((((((.(.(..((.(((....))).))...).)------------.))))))).)))))))))))...........((((....)))))))))....... ( -41.90) >DroSim_CAF1 25 120 + 1 AUCGCCUUAGGGUUGUUUUGGCAGCGGGUGCUACCAGGAGACUGCUACCAGGCCCGGGUUGCCCCGCUCGCCAUAAACGAUCCUGUAAUUCUCGUUGGGCGCGCGCUCGCGAUUGUUCUU (((((..(((((((((((((((((((((.((.(((.((.(.(((....))).))).))).)))))))).)))).)))))))))))...........((((....)))))))))....... ( -54.10) >DroYak_CAF1 21 117 + 1 AUCGCCUUAGGGUUGUUUUGGUAGCGGGUGCUACCAGGAGACUACUACCAGGC---GGUUGCCCCGUUCACCAUAAACGAUCCUGUAAUUCUCGUUGGGCGCGCGCUCGCGAUUGUUCUU (((((..(((((((((((((((((((((.((.(((((....)).(.....)..---))).)))))))).)))).)))))))))))...........((((....)))))))))....... ( -46.10) >consensus AUCGCCUUAGGGUUGUUUUGGCAGCGGGUGCUACCAGGAGACUGCUACCAGGC___GGUUGCCCCGCUCGCCAUAAACGAUCCUGUAAUUCUCGUUGGGCGCGCGCUCGCGAUUGUUCUU (((((..(((((((((((((((((((((.((.(((((....))(((....)))...))).)))))))).)))).)))))))))))...........((((....)))))))))....... (-37.59 = -40.37 + 2.78)


| Location | 1,243,852 – 1,243,948 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.32 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -32.89 |
| Energy contribution | -33.03 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1243852 96 - 20766785 AUGCUC------------CUGCUGCCUGGGUGCCCACAUAAAGAACAAUCGCGAGCGCGCGCCCAACGAGAAUUACAGGAUCGUUUAUGGCAACC------------GCCUGGUAGCAGU ......------------((((((((.((((((.........((....))((....))))))))(((((...........)))))...(((....------------))).)))))))). ( -32.60) >DroSim_CAF1 65 108 - 1 AUGCUC------------CUGCUGCCUGGGUGCCCACAUGAAGAACAAUCGCGAGCGCGCGCCCAACGAGAAUUACAGGAUCGUUUAUGGCGAGCGGGGCAACCCGGGCCUGGUAGCAGU .(((((------------(....((((((((((((..............(....)(((.((((.(((((...........)))))...)))).))))))).))))))))..)).)))).. ( -47.30) >DroYak_CAF1 61 117 - 1 AUGCUCCUGCUCCUGCUCCUGCUGCCUGGGUGCUCACAUAAAGAACAAUCGCGAGCGCGCGCCCAACGAGAAUUACAGGAUCGUUUAUGGUGAACGGGGCAACC---GCCUGGUAGUAGU ..((....))........(((((((((((((((.........((....))((....)))))))))...........(((..(((((.....)))))((....))---.))))))))))). ( -37.10) >consensus AUGCUC____________CUGCUGCCUGGGUGCCCACAUAAAGAACAAUCGCGAGCGCGCGCCCAACGAGAAUUACAGGAUCGUUUAUGGCGAACGGGGCAACC___GCCUGGUAGCAGU ..................((((((((.((((((.........((....))((....))))))))(((((...........)))))...(((....(((....)))..))).)))))))). (-32.89 = -33.03 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:02 2006