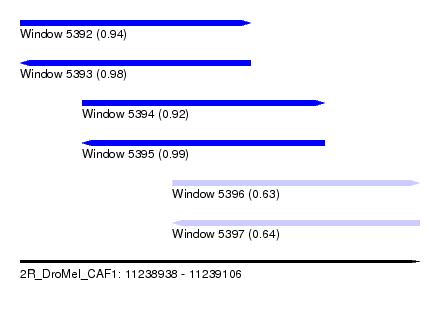
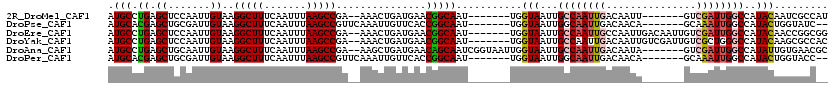
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,238,938 – 11,239,106 |
| Length | 168 |
| Max. P | 0.992694 |

| Location | 11,238,938 – 11,239,035 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 68.40 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -11.71 |
| Energy contribution | -12.97 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.36 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939843 |
| Prediction | RNA |
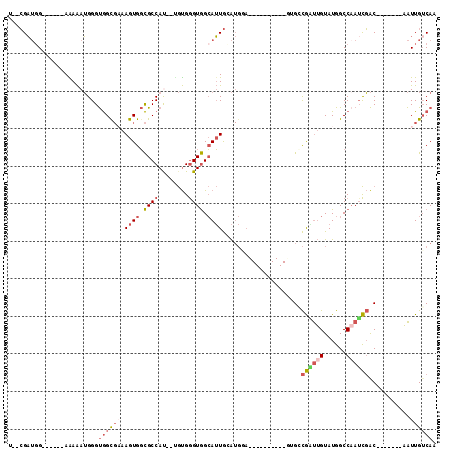
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11238938 97 + 20766785 U--CGAUUG------AAAAAUGGGUGGCGCAAGUGGCGCCAU--UGUGGGUGGCAUUGAAUGGAUGCAGGUAGUAUGGCGAUUGUAUGGCCAAUCGAC-------AAUUGUCAA .--(.(((.------...))).)(((((((.....)))))))--........(((((.....)))))........((((((((((.(((....)))))-------)))))))). ( -28.40) >DroSec_CAF1 25613 97 + 1 U--CGAUGG------AGAAAUGGGUGGCGAAAGUGGCGCCAU--CGUGGGUGGCAUUGGAUGGAUGCAGGUAGUGUGCCGAUUGUAUGGCCAAUCGAC-------AAUUGUCAA .--((((((------(....)..((.((....)).)).))))--))..((((.(((((..((....))..)))))))))(((((......)))))(((-------....))).. ( -28.50) >DroSim_CAF1 28241 97 + 1 U--CGAUGG------AGAAAUGGGUGGCGAAAGUGGCGCCAU--UGUGGGUGGCAUUGCAUGGAUGCAGGUAGUGCGCCGAUUGUAUGGCCAAUCGAC-------AAUUGUCAA .--.((..(------...(((((((.((....)).)).))))--)((.((((((..(((((((.((((.....)))))))..))))..))).))).))-------..)..)).. ( -33.00) >DroEre_CAF1 25923 94 + 1 U--CGAUGG------AAAAAUGGGUGGUGAAAGUGGCGCCAU--UGUGGGUGGCAUUGCAUGCA----------CCGCCGGUUGUAUGGCCAAUCGACAAUUGUCAAUUGGCAA .--......------...(((.(((((((..((((.((((..--....)))).)))).....))----------))))).))).....((((((.(((....))).)))))).. ( -38.00) >DroYak_CAF1 27005 96 + 1 UGGCGAUGG------AAAAAUGGGUGGUGAAAGUGGCGCCAC--AGUGGGUGGCAUUGGAUGCA----------GUGGCGCUUGUAUGGCCCAGCGACAAUCGACAAUUGUCAA (((((((..------.....(((((.(((.(((((.((((((--.....)))))((((....))----------))).))))).))).)))))(((.....)).).))))))). ( -34.70) >DroPer_CAF1 27375 87 + 1 C--AGCAGGAGGCCCAGAGAGGGGCAGC--CAGUGCUGCCACAAUACGAGUAGGAGUACG----------------GGUACCAGUAUGGCCAAUUUGC-------UGUUGUCAA (--((((((..((((......))))..)--)..)))))..(((((((((((.((.((((.----------------(....).))))..)).))))).-------))))))... ( -30.20) >consensus U__CGAUGG______AAAAAUGGGUGGCGAAAGUGGCGCCAU__UGUGGGUGGCAUUGCAUGGA__________GUGCCGAUUGUAUGGCCAAUCGAC_______AAUUGUCAA ........................((((((.((((.((((........)))).)))).....................((((((......))))))...........)))))). (-11.71 = -12.97 + 1.25)

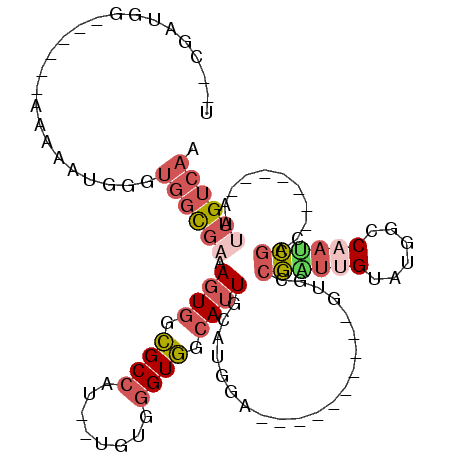
| Location | 11,238,938 – 11,239,035 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 68.40 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -7.93 |
| Energy contribution | -8.07 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.33 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11238938 97 - 20766785 UUGACAAUU-------GUCGAUUGGCCAUACAAUCGCCAUACUACCUGCAUCCAUUCAAUGCCACCCACA--AUGGCGCCACUUGCGCCACCCAUUUUU------CAAUCG--A ((((..(((-------((((((((......))))))...........((((.......)))).....)))--))(((((.....))))).........)------)))...--. ( -20.70) >DroSec_CAF1 25613 97 - 1 UUGACAAUU-------GUCGAUUGGCCAUACAAUCGGCACACUACCUGCAUCCAUCCAAUGCCACCCACG--AUGGCGCCACUUUCGCCACCCAUUUCU------CCAUCG--A ........(-------((((((((......)))))))))........((((.......))))......((--((((.(...................).------))))))--. ( -20.11) >DroSim_CAF1 28241 97 - 1 UUGACAAUU-------GUCGAUUGGCCAUACAAUCGGCGCACUACCUGCAUCCAUGCAAUGCCACCCACA--AUGGCGCCACUUUCGCCACCCAUUUCU------CCAUCG--A ..(((....-------)))(((((......)))))((((.......((((....))))..((((......--.))))........))))..........------......--. ( -22.40) >DroEre_CAF1 25923 94 - 1 UUGCCAAUUGACAAUUGUCGAUUGGCCAUACAACCGGCGG----------UGCAUGCAAUGCCACCCACA--AUGGCGCCACUUUCACCACCCAUUUUU------CCAUCG--A ..((((((((((....)))))))))).........((.((----------((.......(((((......--.))))).......)))).)).......------......--. ( -28.34) >DroYak_CAF1 27005 96 - 1 UUGACAAUUGUCGAUUGUCGCUGGGCCAUACAAGCGCCAC----------UGCAUCCAAUGCCACCCACU--GUGGCGCCACUUUCACCACCCAUUUUU------CCAUCGCCA ..(((((((...)))))))..((((........(((((((----------.((((...))))........--)))))))...........)))).....------......... ( -22.21) >DroPer_CAF1 27375 87 - 1 UUGACAACA-------GCAAAUUGGCCAUACUGGUACC----------------CGUACUCCUACUCGUAUUGUGGCAGCACUG--GCUGCCCCUCUCUGGGCCUCCUGCU--G .......((-------(((....((((((((.((((..----------------........)))).))))...((((((....--))))))........))))...))))--) ( -28.20) >consensus UUGACAAUU_______GUCGAUUGGCCAUACAAUCGCCAC__________UCCAUGCAAUGCCACCCACA__AUGGCGCCACUUUCGCCACCCAUUUCU______CCAUCG__A ................(.((((((......)))))).).....................(((((.........))))).................................... ( -7.93 = -8.07 + 0.14)
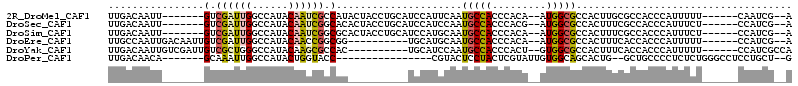
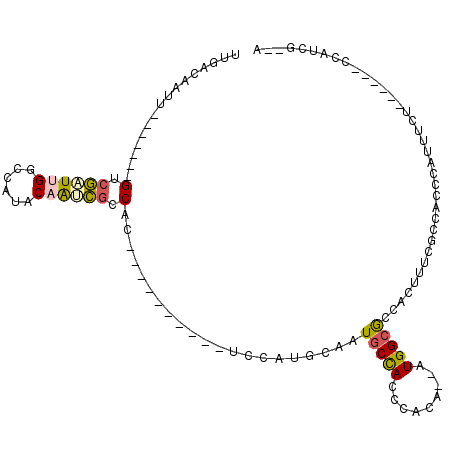
| Location | 11,238,964 – 11,239,066 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.73 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -16.65 |
| Energy contribution | -17.52 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11238964 102 + 20766785 GGCGCCAU--UGUGGGUGGCAUUGAAUGGAUGCAGGUAGUAUGGCGAUUGUAUGGCCAAUCGAC-------AAUUGUCAAUUGGCAAUUACCAAUUGCCGUUCAUCAGUUU-- ..((((..--....))))(((((.....))))).(((.(.((((((((((((..((((((.(((-------....))).))))))...)).)))))))))).)))).....-- ( -36.30) >DroSec_CAF1 25639 102 + 1 GGCGCCAU--CGUGGGUGGCAUUGGAUGGAUGCAGGUAGUGUGCCGAUUGUAUGGCCAAUCGAC-------AAUUGUCAAUUGGCAAUUACCAAUUGCCGUUCAUCAGUUU-- ...(((((--.....)))))....((((((((((((((((.(((((((((......)))))(((-------....)))....))))))))))...)).)))))))).....-- ( -37.90) >DroSim_CAF1 28267 102 + 1 GGCGCCAU--UGUGGGUGGCAUUGCAUGGAUGCAGGUAGUGCGCCGAUUGUAUGGCCAAUCGAC-------AAUUGUCAAUUGGCAAUUACCAAUUGCCGUUCAUCAGUUU-- ......((--((((..(((((.((((....))))((((((..((((((((......)))))(((-------....)))....))).))))))...)))))..)).))))..-- ( -36.30) >DroEre_CAF1 25949 99 + 1 GGCGCCAU--UGUGGGUGGCAUUGCAUGCA----------CCGCCGGUUGUAUGGCCAAUCGACAAUUGUCAAUUGGCAAUUGGCAAUUACCAAUUGCCGUUCAUCAGUUU-- ((((((((--.....)))))....((((((----------((...)).)))))))))....(((....)))....(((((((((......)))))))))............-- ( -36.30) >DroYak_CAF1 27033 99 + 1 GGCGCCAC--AGUGGGUGGCAUUGGAUGCA----------GUGGCGCUUGUAUGGCCCAGCGACAAUCGACAAUUGUCAAUUGGCAAUUACCAAUUGCCGUUCAUCAGUUU-- ((.((((.--.(..((((.(((((....))----------))).))))..).))))))(((((((((.....))))))....((((((.....))))))))).........-- ( -35.20) >DroPer_CAF1 27405 90 + 1 GCUGCCACAAUACGAGUAGGAGUACG----------------GGUACCAGUAUGGCCAAUUUGC-------UGUUGUCAAUUGCCAAUUACCAAUUGCCGGUGAACAAUUUGA ...((((((((((((((.((.((((.----------------(....).))))..)).))))).-------))))))((((((........))))))..)))........... ( -19.70) >consensus GGCGCCAU__UGUGGGUGGCAUUGCAUGGA__________GUGCCGAUUGUAUGGCCAAUCGAC_______AAUUGUCAAUUGGCAAUUACCAAUUGCCGUUCAUCAGUUU__ ((((((((.......))))).............................(((..((((((.(((...........))).))))))...))).....))).............. (-16.65 = -17.52 + 0.86)
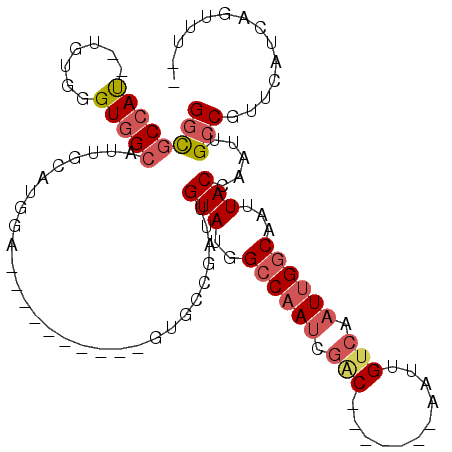
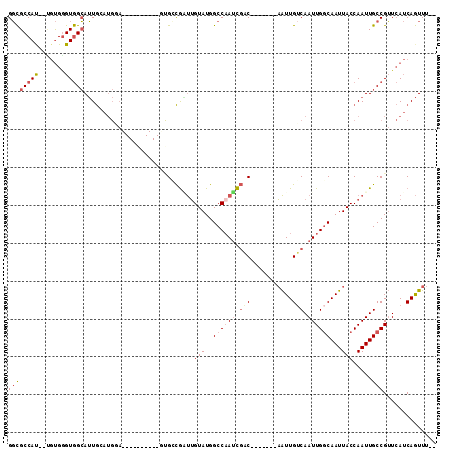
| Location | 11,238,964 – 11,239,066 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.73 |
| Mean single sequence MFE | -29.85 |
| Consensus MFE | -16.19 |
| Energy contribution | -16.50 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11238964 102 - 20766785 --AAACUGAUGAACGGCAAUUGGUAAUUGCCAAUUGACAAUU-------GUCGAUUGGCCAUACAAUCGCCAUACUACCUGCAUCCAUUCAAUGCCACCCACA--AUGGCGCC --............(((.....(((...((((((((((....-------))))))))))..)))....(((((.......((((.......))))........--)))))))) ( -29.56) >DroSec_CAF1 25639 102 - 1 --AAACUGAUGAACGGCAAUUGGUAAUUGCCAAUUGACAAUU-------GUCGAUUGGCCAUACAAUCGGCACACUACCUGCAUCCAUCCAAUGCCACCCACG--AUGGCGCC --.....((((...((((((((((....)))))))).....(-------((((((((......))))))))).....))..))))........((((......--.))))... ( -30.80) >DroSim_CAF1 28267 102 - 1 --AAACUGAUGAACGGCAAUUGGUAAUUGCCAAUUGACAAUU-------GUCGAUUGGCCAUACAAUCGGCGCACUACCUGCAUCCAUGCAAUGCCACCCACA--AUGGCGCC --............(((....((((..(((((((((((....-------))))))))(((........)))))).))))((((....))))..((((......--.))))))) ( -31.20) >DroEre_CAF1 25949 99 - 1 --AAACUGAUGAACGGCAAUUGGUAAUUGCCAAUUGCCAAUUGACAAUUGUCGAUUGGCCAUACAACCGGCGG----------UGCAUGCAAUGCCACCCACA--AUGGCGCC --............((((((((((....))))))))))(((((((....))))))).(((((......((((.----------((....)).)))).......--)))))... ( -35.42) >DroYak_CAF1 27033 99 - 1 --AAACUGAUGAACGGCAAUUGGUAAUUGCCAAUUGACAAUUGUCGAUUGUCGCUGGGCCAUACAAGCGCCAC----------UGCAUCCAAUGCCACCCACU--GUGGCGCC --.....((((..(((((((((((....)))))))(((((((...)))))))))))((((......).)))..----------..))))....(((((.....--)))))... ( -32.80) >DroPer_CAF1 27405 90 - 1 UCAAAUUGUUCACCGGCAAUUGGUAAUUGGCAAUUGACAACA-------GCAAAUUGGCCAUACUGGUACC----------------CGUACUCCUACUCGUAUUGUGGCAGC .(((.(((((......((((((.(....).)))))).....)-------)))).)))((((((...((((.----------------.(((....)))..))))))))))... ( -19.30) >consensus __AAACUGAUGAACGGCAAUUGGUAAUUGCCAAUUGACAAUU_______GUCGAUUGGCCAUACAAUCGCCAC__________UCCAUGCAAUGCCACCCACA__AUGGCGCC ..............(.((((((((....)))))))).)...........(.((((((......)))))).).....................(((((.........))))).. (-16.19 = -16.50 + 0.31)
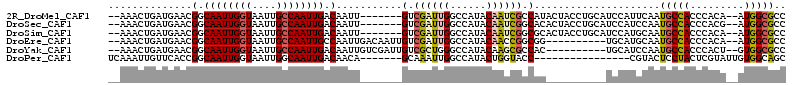
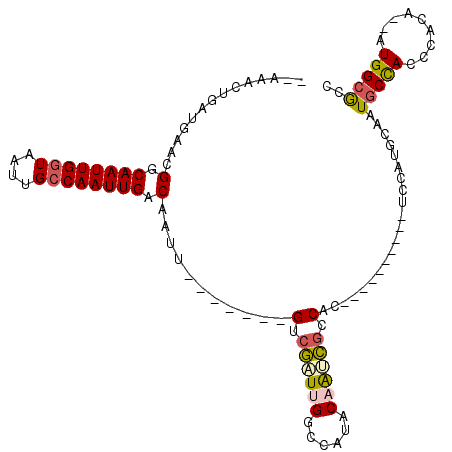
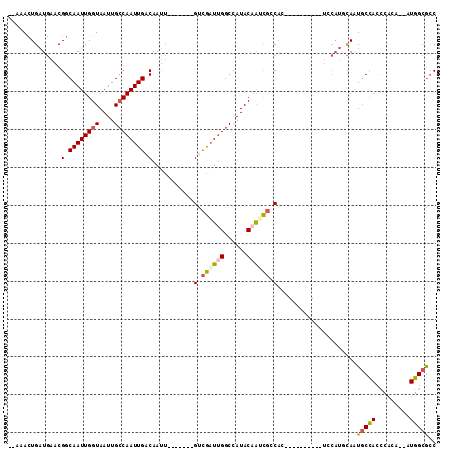
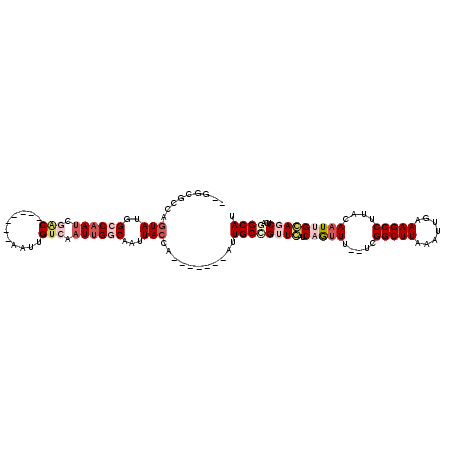
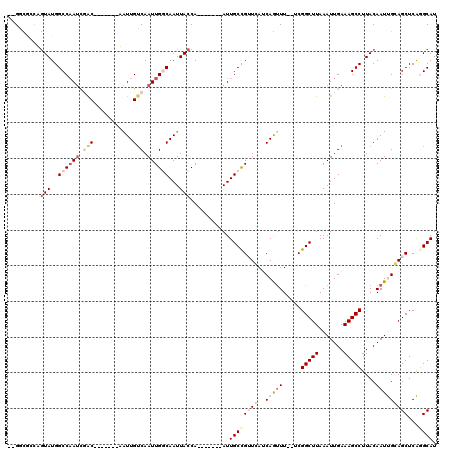
| Location | 11,239,002 – 11,239,106 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.27 |
| Mean single sequence MFE | -30.63 |
| Consensus MFE | -14.19 |
| Energy contribution | -16.05 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11239002 104 + 20766785 AUGGCGAUUGUAUGGCCAAUCGAC-------AAUUGUCAAUUGGCAAUUACCA-------AUUGCCGUUCAUCAGUUU--UCGGCUUAAAUUGAAAGCCUUACAAUUGGAGCUCAGGCAU ((((((((((((..((((((.(((-------....))).))))))...)).))-------))))))))...((((((.--..(((((.......)))))....)))))).((....)).. ( -34.40) >DroPse_CAF1 27922 104 + 1 --GAUACCAGUAUGGCCAAUUUGC-------UGUUGUCAAUUGCCAAUUACCA-------AUUGCCGGUGAACAAUUUGAACGGCUUAAAUUGAAAGCCUUACAAUCGCAGCUCGUGCAU --.......(((((((.....(((-------.(((((((((((........))-------))))..(((...((((((((.....))))))))...)))..))))).))))).))))).. ( -24.30) >DroEre_CAF1 25977 111 + 1 CCGCCGGUUGUAUGGCCAAUCGACAAUUGUCAAUUGGCAAUUGGCAAUUACCA-------AUUGCCGUUCAUCAGUUU--UCGGCUUAAAUUGAAAGCCUUACAAUUGGAGCUCAGGCAU ...(((((((((.(((.....(((....)))....(((((((((......)))-------)))))).....(((((((--.......)))))))..))).))))))))).((....)).. ( -38.60) >DroYak_CAF1 27061 111 + 1 GUGGCGCUUGUAUGGCCCAGCGACAAUCGACAAUUGUCAAUUGGCAAUUACCA-------AUUGCCGUUCAUCAGUUU--UCGGCUUAAAUUGAAAGCCUUACAAUUGGAGCUCAGGCAU .....(((((...(((((((........((.(((((.....(((((((.....-------))))))).....))))).--))(((((.......)))))......)))).)))))))).. ( -31.80) >DroAna_CAF1 24035 111 + 1 GCGUUCACAAUAUGGCCAAUCGAC-------UAUUGUCAAUUGGCAAUUACCAAUUACCGAUUGCUGUUCAUCAGCUU--UCGGCUUAAAUUGAAAGCCUUACAAUUGCAGCUCAGGCAU ((.((((.......((((((.(((-------....))).))))))............((((..((((.....))))..--)))).......)))).))............((....)).. ( -30.40) >DroPer_CAF1 27431 104 + 1 --GGUACCAGUAUGGCCAAUUUGC-------UGUUGUCAAUUGCCAAUUACCA-------AUUGCCGGUGAACAAUUUGAACGGCUUAAAUUGAAAGCCUUACAAUCGCAGCUCGUGCAU --.......(((((((.....(((-------.(((((((((((........))-------))))..(((...((((((((.....))))))))...)))..))))).))))).))))).. ( -24.30) >consensus __GGCGCCAGUAUGGCCAAUCGAC_______AAUUGUCAAUUGGCAAUUACCA_______AUUGCCGUUCAUCAGUUU__UCGGCUUAAAUUGAAAGCCUUACAAUUGCAGCUCAGGCAU .........(((..((((((.(((...........))).))))))...)))...........((((((((..(((((.....(((((.......)))))....)))))))))...)))). (-14.19 = -16.05 + 1.86)

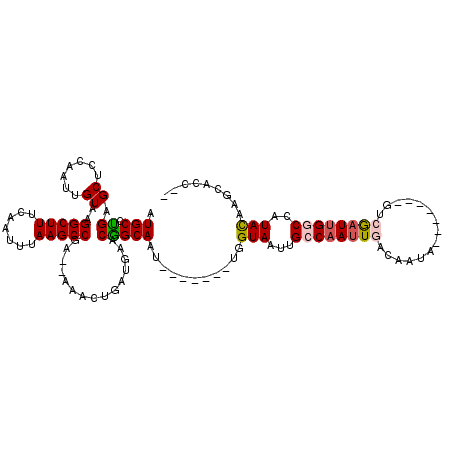
| Location | 11,239,002 – 11,239,106 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.27 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.76 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11239002 104 - 20766785 AUGCCUGAGCUCCAAUUGUAAGGCUUUCAAUUUAAGCCGA--AAACUGAUGAACGGCAAU-------UGGUAAUUGCCAAUUGACAAUU-------GUCGAUUGGCCAUACAAUCGCCAU .((((.(..(..((.((....(((((.......)))))..--.)).))..)..)))))..-------(((..(((((((((((((....-------)))))))))).....)))..))). ( -31.30) >DroPse_CAF1 27922 104 - 1 AUGCACGAGCUGCGAUUGUAAGGCUUUCAAUUUAAGCCGUUCAAAUUGUUCACCGGCAAU-------UGGUAAUUGGCAAUUGACAACA-------GCAAAUUGGCCAUACUGGUAUC-- .(((.((....((((((....(((((.......))))).....))))))....)))))((-------..(((..((((((((..(....-------)..)))).)))))))..))...-- ( -26.30) >DroEre_CAF1 25977 111 - 1 AUGCCUGAGCUCCAAUUGUAAGGCUUUCAAUUUAAGCCGA--AAACUGAUGAACGGCAAU-------UGGUAAUUGCCAAUUGCCAAUUGACAAUUGUCGAUUGGCCAUACAACCGGCGG ..((((..((.......)).))))...........((((.--......(((..(((((((-------((((....))))))))))(((((((....))))))))..))).....)))).. ( -37.40) >DroYak_CAF1 27061 111 - 1 AUGCCUGAGCUCCAAUUGUAAGGCUUUCAAUUUAAGCCGA--AAACUGAUGAACGGCAAU-------UGGUAAUUGCCAAUUGACAAUUGUCGAUUGUCGCUGGGCCAUACAAGCGCCAC ..((((.(((..(((((....(((((.......)))))..--.....((..(....((((-------((((....))))))))....)..)))))))..))))))).............. ( -32.40) >DroAna_CAF1 24035 111 - 1 AUGCCUGAGCUGCAAUUGUAAGGCUUUCAAUUUAAGCCGA--AAGCUGAUGAACAGCAAUCGGUAAUUGGUAAUUGCCAAUUGACAAUA-------GUCGAUUGGCCAUAUUGUGAACGC ..((((..((.......)).))))..((((((...(((((--..((((.....))))..))))))))))).....((((((((((....-------)))))))))).............. ( -35.20) >DroPer_CAF1 27431 104 - 1 AUGCACGAGCUGCGAUUGUAAGGCUUUCAAUUUAAGCCGUUCAAAUUGUUCACCGGCAAU-------UGGUAAUUGGCAAUUGACAACA-------GCAAAUUGGCCAUACUGGUACC-- .(((.((....((((((....(((((.......))))).....))))))....)))))((-------..(((..((((((((..(....-------)..)))).)))))))..))...-- ( -26.30) >consensus AUGCCUGAGCUCCAAUUGUAAGGCUUUCAAUUUAAGCCGA__AAACUGAUGAACGGCAAU_______UGGUAAUUGCCAAUUGACAAUA_______GUCGAUUGGCCAUACAAGCACC__ .(((.((.((.......))..(((((.......)))))...............)))))...........(((...((((((((...............))))))))..)))......... (-14.48 = -14.76 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:04 2006