| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,231,393 – 11,231,492 |
| Length | 99 |
| Max. P | 0.518293 |

| Location | 11,231,393 – 11,231,492 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.21 |
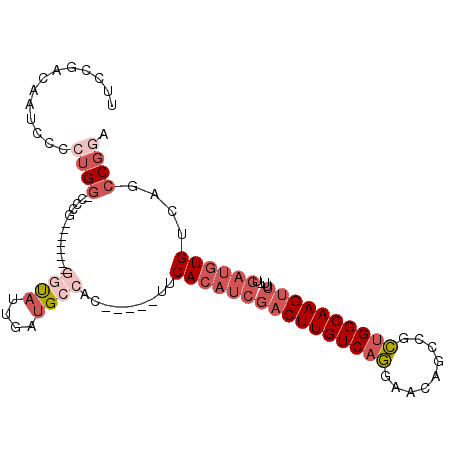
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -15.55 |
| Energy contribution | -17.80 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518293 |
| Prediction | RNA |
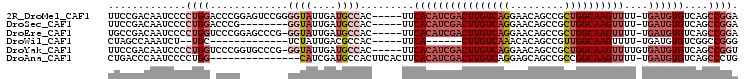
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11231393 99 + 20766785 UUCCGACAAUCCCCUGGACCCGGAGUCCGGGGUAUUGAUGCCAC-----UUCACAUCGACUUGUCAGGAACAGCCGCUGGCAAGUUUU-UGAUGUGUCAGCCGGA .((((.((((.((((((((.....)))))))).))))..((...-----..(((((((((((((((((......).))))))))))..-.))))))...)))))) ( -42.80) >DroSec_CAF1 18360 91 + 1 UUCCGACAAUCCCCUGGACCCG--------GGUAUUGAUGCCAC-----UUCACAUCGACUUGUCAGGAACAGCCGCUGGCAAGUUUU-UGAUGUGUCAGCCGGA .((((.((((.(((.(....))--------)).))))..((...-----..(((((((((((((((((......).))))))))))..-.))))))...)))))) ( -31.90) >DroEre_CAF1 18475 98 + 1 UGCCGACAAUCCCCUGGUCCCGGAGCCCG-GGUAUUGAUGCCAC-----UUCACAUCGACUUGUCAGGAACAGCCGCUGGCAAGUUUU-UGAUGUGUCAGCCGGA ..(((.((((.(((.(((......))).)-)).))))..((...-----..(((((((((((((((((......).))))))))))..-.))))))...))))). ( -35.40) >DroWil_CAF1 39274 78 + 1 CUAGCCAAAUCU--UGC-------------UCUAUUGACGCCAC-----UUCA------CUUGUCAAACACAGCCGUUGGCAAGUUUU-UGAUGUGUCGGCCGGG (..(((......--.((-------------((....)).))(((-----.(((------((((((((.(......))))))))))...-.)).)))..)))..). ( -19.40) >DroYak_CAF1 18670 99 + 1 UUCCGACAAUCCCCUGGUCCCGGUGCCCG-GGUAUUGAUGCCAC-----UUCACAUCGACUUGUCAGGAACAGCCGCUGGCAAGUUUUGUGAUGUGUCAGCCGGU ..(((.((((.(((.(((......))).)-)).)))).((.(((-----.(((((..(((((((((((......).)))))))))).))))).))).))..))). ( -37.90) >DroAna_CAF1 17460 89 + 1 CUGACCCAAUCCCCUGG---------------CAUCGAUGCCACUUCACUUCACAUCGACUUGUCAGGAGCAGCCGCCGGCAAGUUUU-UGAUGUGUCAGCCCUG (((((.........(((---------------((....))))).........(((((((((((((.((........))))))))))..-.))))))))))..... ( -25.90) >consensus UUCCGACAAUCCCCUGG_CCCG________GGUAUUGAUGCCAC_____UUCACAUCGACUUGUCAGGAACAGCCGCUGGCAAGUUUU_UGAUGUGUCAGCCGGA .............((((.............((((....)))).........((((((((((((((((.........))))))))))....))))))....)))). (-15.55 = -17.80 + 2.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:57 2006