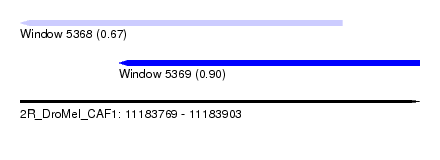
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,183,769 – 11,183,903 |
| Length | 134 |
| Max. P | 0.903370 |

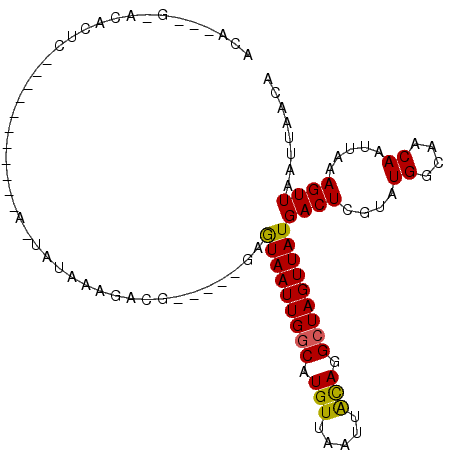
| Location | 11,183,769 – 11,183,877 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -15.05 |
| Energy contribution | -14.92 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11183769 108 - 20766785 AUACAGAUG-----GAGUAAUUGGCAUGUUAAUUACAGGCUAGUUAUGACUCGUAUGGCAACAAUUAAAGUUAAUUAACAUUCGAAA-ACAA-ACUG--CAAA-AAGCCGUCUAAAUC ....(((((-----(.(((((((((.(((.....))).)))))))))...(((..((....))......(((....)))...)))..-....-....--....-...))))))..... ( -22.00) >DroPse_CAF1 98361 108 - 1 ACAAAGGCG-----CAGUAAUUGGCAUGUUAAUUGCAGGCUAGUUAUGACUCGUAUGGCAACAAUUAAAGUUAAUUAACAUUCGAUC-ACAA-ACUG--CAAA-CAGUCGCCAGAGUG .....((((-----..((.(((((.(((((((((((..((.(((....))).)).((....))......).))))))))))))))).-))..-((((--....-))))))))...... ( -24.60) >DroWil_CAF1 145342 103 - 1 AUAAAGACAACGACAAAUAAUUGGCAUGUUAAUUAUAGGAUAGUUAUGACUCGAAUGGCAACAAUUAAAGUUAAUUAACAUUCGAAU-GUUGCUCUGUC--------------GGCUU .....((((.(((((.((((((((....)))))))).((((.(((((((((..((((....).)))..)))))..))))))))...)-))))...))))--------------..... ( -21.00) >DroYak_CAF1 98395 109 - 1 AUACAGAUG-----AAGUAAUUGGCAUGUUAAUUACGGGCUAGUUAUGACUCGUAUGGCAACAAUUAAAGUUAAUUAACAUUCGAAA-ACAA-ACUG--CAAAAAAGCCGUCUAAAUG ....(((((-----..((..((((.(((((((((((((((.......).))))).((....)).........)))))))))))))..-))..-...(--(......)))))))..... ( -20.50) >DroAna_CAF1 105925 108 - 1 AGCCAUGCG-----GUGUAAUUGGCAUGUUAAUUACAGGCUAGUUAUGACUCGUAUGGCAACAAUUAAAGUUAAUUAACAUUCGCAAAUCAA-ACUG---AAA-AAGUCGCCCGAGUG .((((((((-----(((((((((((.(((.....))).))))))))).)).))))))))...................((((((........-(((.---...-.)))....)))))) ( -31.20) >DroPer_CAF1 98136 108 - 1 ACAAAGGCG-----CAGUAAUUGGCAUGUUAAUUGCAGGCUAGUUAUGACUCGUAUGGCAACAAUUAAAGUUAAUUAACAUUCGAUC-ACAA-ACUG--CAAA-CAGUCGCCAGAGUG .....((((-----..((.(((((.(((((((((((..((.(((....))).)).((....))......).))))))))))))))).-))..-((((--....-))))))))...... ( -24.60) >consensus AUAAAGACG_____AAGUAAUUGGCAUGUUAAUUACAGGCUAGUUAUGACUCGUAUGGCAACAAUUAAAGUUAAUUAACAUUCGAAA_ACAA_ACUG__CAAA_AAGUCGCCAGAGUG .....((((.......(((((((((.(((.....))).)))))))))((((....((....)).....))))....................................))))...... (-15.05 = -14.92 + -0.14)

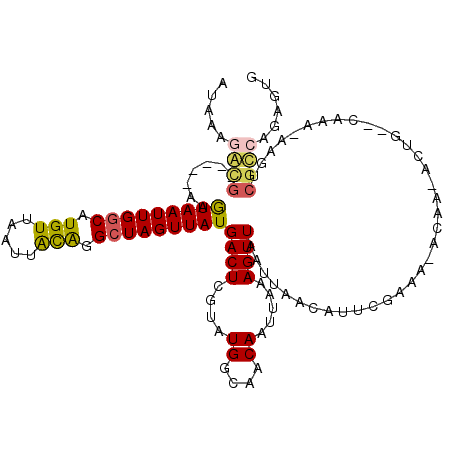
| Location | 11,183,802 – 11,183,903 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -19.23 |
| Consensus MFE | -13.23 |
| Energy contribution | -12.90 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11183802 101 - 20766785 ACACUCGCACACUC-GCUGGCUUACA-UAUACAGAUG-----GAGUAAUUGGCAUGUUAAUUACAGGCUAGUUAUGACUCGUAUGGCAACAAUUAAAGUUAAUUAACA ......((......-))((((((...-.((((.((.(-----..(((((((((.(((.....))).)))))))))..)))))))(....).....))))))....... ( -21.00) >DroPse_CAF1 98394 86 - 1 ACAU--GUACA---------------GCACAAAGGCG-----CAGUAAUUGGCAUGUUAAUUGCAGGCUAGUUAUGACUCGUAUGGCAACAAUUAAAGUUAAUUAACA ....--.....---------------((.((...(((-----..(((((((((.(((.....))).)))))))))....))).))))..................... ( -17.00) >DroSim_CAF1 97389 98 - 1 ACACUCGCACACUCCGCC-----ACACUAUACAGAUG-----GAGUAAUUGGCAUGUUAAUUACAGGCUAGUUAUGACUCGUAUGGCAACAAUUAAAGUUAAUUAACA ...............(((-----(.....(((.((.(-----..(((((((((.(((.....))).)))))))))..))))))))))..................... ( -20.00) >DroWil_CAF1 145365 87 - 1 ----------ACUC-----------ACUAUAAAGACAACGACAAAUAAUUGGCAUGUUAAUUAUAGGAUAGUUAUGACUCGAAUGGCAACAAUUAAAGUUAAUUAACA ----------..((-----------.((((((.((((....(((....)))...))))..))))))))..(((((((((..((((....).)))..)))))..)))). ( -13.30) >DroAna_CAF1 105958 92 - 1 ---------CACUC-ACUCAUAUGUA-UAGCCAUGCG-----GUGUAAUUGGCAUGUUAAUUACAGGCUAGUUAUGACUCGUAUGGCAACAAUUAAAGUUAAUUAACA ---------.....-...........-..((((((((-----(((((((((((.(((.....))).))))))))).)).))))))))..................... ( -27.10) >DroPer_CAF1 98169 86 - 1 ACAU--GUACA---------------GCACAAAGGCG-----CAGUAAUUGGCAUGUUAAUUGCAGGCUAGUUAUGACUCGUAUGGCAACAAUUAAAGUUAAUUAACA ....--.....---------------((.((...(((-----..(((((((((.(((.....))).)))))))))....))).))))..................... ( -17.00) >consensus ACA___G_ACACUC___________A_UAUAAAGACG_____GAGUAAUUGGCAUGUUAAUUACAGGCUAGUUAUGACUCGUAUGGCAACAAUUAAAGUUAAUUAACA ............................................(((((((((.(((.....))).)))))))))((((....((....)).....))))........ (-13.23 = -12.90 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:37 2006