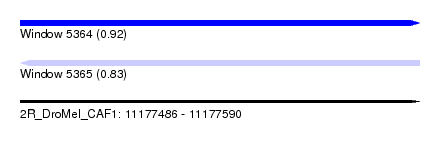
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,177,486 – 11,177,590 |
| Length | 104 |
| Max. P | 0.918696 |

| Location | 11,177,486 – 11,177,590 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -47.80 |
| Consensus MFE | -26.76 |
| Energy contribution | -28.93 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
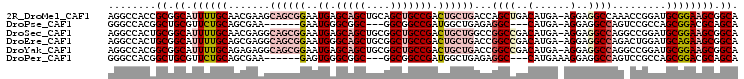
>2R_DroMel_CAF1 11177486 104 + 20766785 AGGCCACCGCGGCAUUUUGCAACGAAGCAGCGGAAUGAGCAGCUGCAGCUGCCGACUGCUGACCAGCUGACAUGA-AGGAGGCCAAACCGGAUGCGGAAGCGGCA .((((.((((..((((((((.........)))))))).))(((((((((........))))..))))).......-.)).))))........(((....)))... ( -39.30) >DroPse_CAF1 92356 92 + 1 GGGCCACGGCUGCGUUCUGCAGCGAA------GAAUGGGCGGC---GGCGGCCGAUGGCUGAGAGGC---CAUGA-AGGAGGCCAGUCCGCCAGCGGACGCAGCA ........(((((((.((((.((...------......))(((---(((((((.((((((....)))---)))..-....)))).).))))).))))))))))). ( -48.60) >DroSec_CAF1 92053 104 + 1 AGGCCACUGCGGCAUUUUGCAACGAGGCAGCGGAAUGAGCAGCUGCGGCUGCCGACUGCUGGCCGGCCGACAUGA-AGGAGGCCAGGCCGGAUGCGGAAGCGGCA ..(((.((((..((((((((.........)))))))).)))).....(((.(((....((((((((((..(....-.)..)))).))))))...))).)))))). ( -50.70) >DroEre_CAF1 94604 104 + 1 AGGCCACUGCGGCAUUUUGCAGCGAGGCAGCGGAAUGGGCAGCUGCGGCUGCCGACUGCUGACCGGCCGACAUGA-AGGAGGCCAGACUGGAUGCAGAAGCGGCA ..(((.((...(((((...((((..((((((((..(.((((((....)))))).))))))).))((((..(....-.)..)))).).))))))))...)).))). ( -49.40) >DroYak_CAF1 92104 104 + 1 AGGCCACGGCGGCAUUUUGCAGAGAGGCAGCGGAAUGAGCAGCUGCGGCUGCCGACUGCUGACCGGCCGACAUGA-AGGAGGCCAGGCCGGAUGCGGAAGCGGCA ..(((.((((.((.....)).....((((((((..((.(((((....))))))).)))))).))((((..(....-.)..))))..))))...((....))))). ( -49.80) >DroPer_CAF1 92149 93 + 1 GGGCCACGGCUGCGUUCUGCAGCGAA------GAGUGGGCGGC---GGCGGCCGAUGGCUGAGAGGC---CAUGAAAGGAGGCCAGUCCGCCAGCGGACGCAGCA ........(((((((.((((.((...------..)).((((((---(((..((.((((((....)))---)))....))..))).).))))).))))))))))). ( -49.00) >consensus AGGCCACGGCGGCAUUUUGCAGCGAAGCAGCGGAAUGAGCAGCUGCGGCUGCCGACUGCUGACCGGCCGACAUGA_AGGAGGCCAGACCGGAUGCGGAAGCGGCA ........((.((.((((((.......((((((..((.(((((....))))))).))))))...((((..(......)..)))).........)))))))).)). (-26.76 = -28.93 + 2.17)
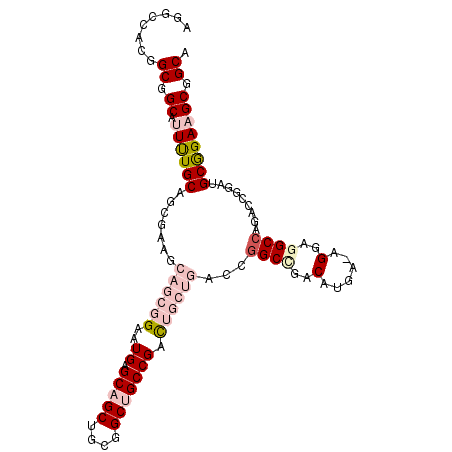

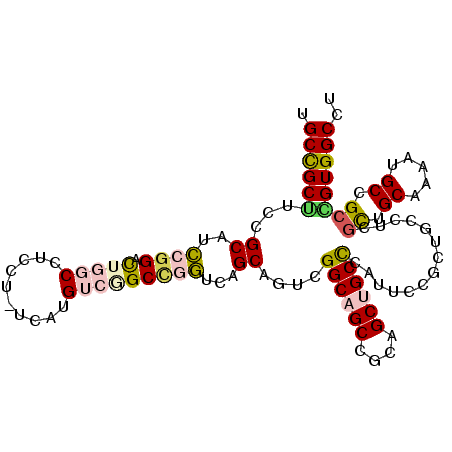
| Location | 11,177,486 – 11,177,590 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -25.61 |
| Energy contribution | -25.92 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11177486 104 - 20766785 UGCCGCUUCCGCAUCCGGUUUGGCCUCCU-UCAUGUCAGCUGGUCAGCAGUCGGCAGCUGCAGCUGCUCAUUCCGCUGCUUCGUUGCAAAAUGCCGCGGUGGCCU ....(((.((((..((((((.(((.....-....)))))))))...(((....(((((.(((((..........)))))...)))))....))).)))).))).. ( -37.50) >DroPse_CAF1 92356 92 - 1 UGCUGCGUCCGCUGGCGGACUGGCCUCCU-UCAUG---GCCUCUCAGCCAUCGGCCGCC---GCCGCCCAUUC------UUCGCUGCAGAACGCAGCCGUGGCCC ....(((.(((.((((.((..((((....-....)---)))..)).)))).))).))).---(((((......------...(((((.....))))).))))).. ( -40.20) >DroSec_CAF1 92053 104 - 1 UGCCGCUUCCGCAUCCGGCCUGGCCUCCU-UCAUGUCGGCCGGCCAGCAGUCGGCAGCCGCAGCUGCUCAUUCCGCUGCCUCGUUGCAAAAUGCCGCAGUGGCCU .((((((...((....((((.((((..(.-....)..)))))))).(((....(((((.(((((..........)))))...)))))....))).)))))))).. ( -44.90) >DroEre_CAF1 94604 104 - 1 UGCCGCUUCUGCAUCCAGUCUGGCCUCCU-UCAUGUCGGCCGGUCAGCAGUCGGCAGCCGCAGCUGCCCAUUCCGCUGCCUCGCUGCAAAAUGCCGCAGUGGCCU .((((((.(.((((...(.((((((..(.-....)..)))))).).(((((.((((((.((....)).......))))))..)))))...)))).).)))))).. ( -44.10) >DroYak_CAF1 92104 104 - 1 UGCCGCUUCCGCAUCCGGCCUGGCCUCCU-UCAUGUCGGCCGGUCAGCAGUCGGCAGCCGCAGCUGCUCAUUCCGCUGCCUCUCUGCAAAAUGCCGCCGUGGCCU .(((((..(.((((..((((.((((..(.-....)..)))))))).((((..((((((.((....)).......))))))...))))...)))).)..))))).. ( -42.30) >DroPer_CAF1 92149 93 - 1 UGCUGCGUCCGCUGGCGGACUGGCCUCCUUUCAUG---GCCUCUCAGCCAUCGGCCGCC---GCCGCCCACUC------UUCGCUGCAGAACGCAGCCGUGGCCC .((.(((.(((.((((.((..((((.........)---)))..)).)))).))).))).---))...((((..------...(((((.....))))).))))... ( -40.50) >consensus UGCCGCUUCCGCAUCCGGACUGGCCUCCU_UCAUGUCGGCCGGUCAGCAGUCGGCAGCCGCAGCUGCCCAUUCCGCUGCCUCGCUGCAAAAUGCCGCCGUGGCCU .((((((...((..((((.(((((..........)))))))))...))....((((((....))))))..............((.((.....)).)))))))).. (-25.61 = -25.92 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:33 2006