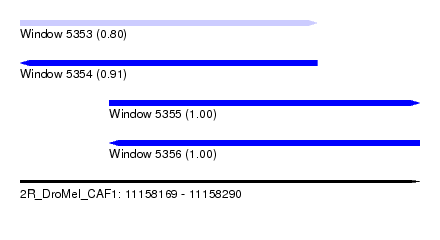
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,158,169 – 11,158,290 |
| Length | 121 |
| Max. P | 0.999638 |

| Location | 11,158,169 – 11,158,259 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -21.26 |
| Energy contribution | -22.34 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11158169 90 + 20766785 UGCCCAUACAACUGGCGGUGGUGAUG---------GUGCUGUUGCAAUUGCUACUGC---UGCAUCUGCAGCUGCAACAGUCAGCGGCGACAUGUUGCGACA .(((.........))).((.(..(((---------.(((((((((..((((....((---(((....))))).))))..).))))))))...)))..).)). ( -33.80) >DroSec_CAF1 72940 84 + 1 UGCCCAUACAACUGGCGGUG---------------GUGCAGUUGCAAUUGCUACUGC---UGCAUCUGCAGCUGCAACAGUCAGCGGCGACAUGUUGCGACA .(((.........)))..((---------------.((((((((((..(((......---.)))..)))))))))).))(((.(((((.....)))))))). ( -33.20) >DroSim_CAF1 72536 84 + 1 UGCCCAUACAACUGGCGGUG---------------GUGCAGUUGCAAUUGCUACUGC---UGCAUCUGCAGCUGCAACAGUCAGCGGCGACAUGUUGCGACA .(((.........)))..((---------------.((((((((((..(((......---.)))..)))))))))).))(((.(((((.....)))))))). ( -33.20) >DroEre_CAF1 75584 69 + 1 UGCCCAUACAAC------------------------------UGCAAUUGCUACUGC---UGCAUCUGCAGCUGCAACAGUCAGCGGCGACAUGUUGCGACA ...........(------------------------------((((..(((......---.)))..))))).((((((((((......))).)))))))... ( -22.20) >DroYak_CAF1 72612 102 + 1 UGCCCAUACAACUGGCGGCGAUGGUGGCAACGCUGGUGGCACUGCAAUUGCUACUGCUACUGCAUCUGCAGCUGCAACAGUCAGCGGCGACAUGUUGCGACA .............(((.(((((((((....))).(((((((..((....))...))))))).)))).)).)))(((((((((......))).)))))).... ( -36.40) >consensus UGCCCAUACAACUGGCGGUG_______________GUGCAGUUGCAAUUGCUACUGC___UGCAUCUGCAGCUGCAACAGUCAGCGGCGACAUGUUGCGACA .(((.........)))........................((((((..(((..........)))..))))))((((((((((......))).)))))))... (-21.26 = -22.34 + 1.08)


| Location | 11,158,169 – 11,158,259 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -23.11 |
| Energy contribution | -23.27 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11158169 90 - 20766785 UGUCGCAACAUGUCGCCGCUGACUGUUGCAGCUGCAGAUGCA---GCAGUAGCAAUUGCAACAGCAC---------CAUCACCACCGCCAGUUGUAUGGGCA ....((((((.((((....)))))))))).(((((....)))---)).((.((....)).)).((.(---------(((.((.((.....)).)))))))). ( -30.80) >DroSec_CAF1 72940 84 - 1 UGUCGCAACAUGUCGCCGCUGACUGUUGCAGCUGCAGAUGCA---GCAGUAGCAAUUGCAACUGCAC---------------CACCGCCAGUUGUAUGGGCA ....((((((.((((....)))))))))).(((((....)))---))....((...(((((((((..---------------....).))))))))...)). ( -33.70) >DroSim_CAF1 72536 84 - 1 UGUCGCAACAUGUCGCCGCUGACUGUUGCAGCUGCAGAUGCA---GCAGUAGCAAUUGCAACUGCAC---------------CACCGCCAGUUGUAUGGGCA ....((((((.((((....)))))))))).(((((....)))---))....((...(((((((((..---------------....).))))))))...)). ( -33.70) >DroEre_CAF1 75584 69 - 1 UGUCGCAACAUGUCGCCGCUGACUGUUGCAGCUGCAGAUGCA---GCAGUAGCAAUUGCA------------------------------GUUGUAUGGGCA .(((((((((.((((....))))))))))((((((((.(((.---......))).)))))------------------------------))).....))). ( -27.70) >DroYak_CAF1 72612 102 - 1 UGUCGCAACAUGUCGCCGCUGACUGUUGCAGCUGCAGAUGCAGUAGCAGUAGCAAUUGCAGUGCCACCAGCGUUGCCACCAUCGCCGCCAGUUGUAUGGGCA .((.(((((.((..(.(((((...(((((.(((((..........))))).)))))..))))).)..))..))))).)).......(((.........))). ( -32.70) >consensus UGUCGCAACAUGUCGCCGCUGACUGUUGCAGCUGCAGAUGCA___GCAGUAGCAAUUGCAACUGCAC_______________CACCGCCAGUUGUAUGGGCA .(((((((((.((((....)))))))))).(((((...(((....))))))))...((((((............................))))))..))). (-23.11 = -23.27 + 0.16)

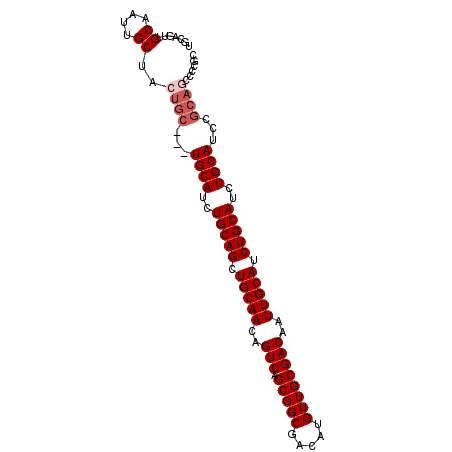
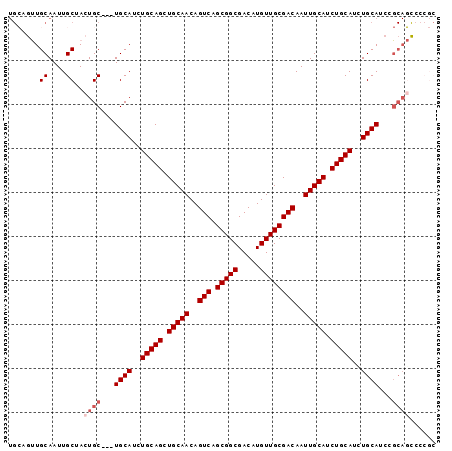
| Location | 11,158,196 – 11,158,290 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 86.80 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -30.84 |
| Energy contribution | -31.84 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.11 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.82 |
| SVM RNA-class probability | 0.999638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11158196 94 + 20766785 UGCUGUUGCAAUUGCUACUGC---UGCAUCUGCAGCUGCAACAGUCAGCGGCGACAUGUUGCGACAAUUGCAUCUGCAUCUGCAUCCGCAGCCCCGC .((((((((((((((....((---(((....))))).(((((((((......))).))))))).))))))))..(((....)))...)))))..... ( -37.30) >DroSec_CAF1 72961 94 + 1 UGCAGUUGCAAUUGCUACUGC---UGCAUCUGCAGCUGCAACAGUCAGCGGCGACAUGUUGCGACAAUUGCAUCUGCAUUUGCAUCCGCAGCCCCGC (((((.(((((((((....((---(((....))))).(((((((((......))).))))))).)))))))).))))).((((....))))...... ( -39.90) >DroSim_CAF1 72557 94 + 1 UGCAGUUGCAAUUGCUACUGC---UGCAUCUGCAGCUGCAACAGUCAGCGGCGACAUGUUGCGACAAUUGCAUCUGCAUCUGCAUCCGCAGCCCCGC (((((.(((((((((....((---(((....))))).(((((((((......))).))))))).)))))))).))))).((((....))))...... ( -41.70) >DroEre_CAF1 75596 75 + 1 ------UGCAAUUGCUACUGC---UGCAUCUGCAGCUGCAACAGUCAGCGGCGACAUGUUGCGACAAUUGCAUCUGCAUCUGCA------------- ------(((((((((....((---(((....))))).(((((((((......))).))))))).))))))))...((....)).------------- ( -30.30) >DroYak_CAF1 72648 92 + 1 UGGCACUGCAAUUGCUACUGCUACUGCAUCUGCAGCUGCAACAGUCAGCGGCGACAUGUUGCGACAAUUGCAUCUGCAUCUGCAGCCGCAUU----- (((((.......))))).(((..(((((..(((((.(((((..(((.(((((.....))))))))..))))).)))))..)))))..)))..----- ( -36.70) >consensus UGCAGUUGCAAUUGCUACUGC___UGCAUCUGCAGCUGCAACAGUCAGCGGCGACAUGUUGCGACAAUUGCAUCUGCAUCUGCAUCCGCAGCCCCGC .......((....))..((((...((((..(((((.(((((..(((.(((((.....))))))))..))))).)))))..))))...))))...... (-30.84 = -31.84 + 1.00)

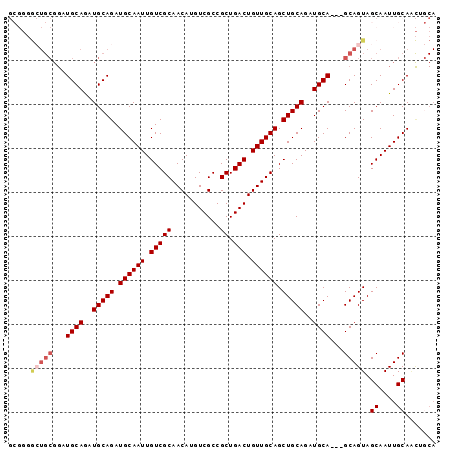
| Location | 11,158,196 – 11,158,290 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 86.80 |
| Mean single sequence MFE | -39.64 |
| Consensus MFE | -31.74 |
| Energy contribution | -32.82 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.93 |
| Structure conservation index | 0.80 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11158196 94 - 20766785 GCGGGGCUGCGGAUGCAGAUGCAGAUGCAAUUGUCGCAACAUGUCGCCGCUGACUGUUGCAGCUGCAGAUGCA---GCAGUAGCAAUUGCAACAGCA (((...((((....)))).))).(.(((((((((.((((((.((((....)))))))))).(((((....)))---))....))))))))).).... ( -40.30) >DroSec_CAF1 72961 94 - 1 GCGGGGCUGCGGAUGCAAAUGCAGAUGCAAUUGUCGCAACAUGUCGCCGCUGACUGUUGCAGCUGCAGAUGCA---GCAGUAGCAAUUGCAACUGCA ((((..(((((........))))).(((((((((.((((((.((((....)))))))))).(((((....)))---))....))))))))).)))). ( -43.10) >DroSim_CAF1 72557 94 - 1 GCGGGGCUGCGGAUGCAGAUGCAGAUGCAAUUGUCGCAACAUGUCGCCGCUGACUGUUGCAGCUGCAGAUGCA---GCAGUAGCAAUUGCAACUGCA ......((((....)))).(((((.(((((((((.((((((.((((....)))))))))).(((((....)))---))....))))))))).))))) ( -43.90) >DroEre_CAF1 75596 75 - 1 -------------UGCAGAUGCAGAUGCAAUUGUCGCAACAUGUCGCCGCUGACUGUUGCAGCUGCAGAUGCA---GCAGUAGCAAUUGCA------ -------------(((....)))..(((((((((.((((((.((((....)))))))))).(((((....)))---))....)))))))))------ ( -32.90) >DroYak_CAF1 72648 92 - 1 -----AAUGCGGCUGCAGAUGCAGAUGCAAUUGUCGCAACAUGUCGCCGCUGACUGUUGCAGCUGCAGAUGCAGUAGCAGUAGCAAUUGCAGUGCCA -----...(((.((((((.(((...(((.......((((((.((((....)))))))))).(((((....))))).)))...))).))))))))).. ( -38.00) >consensus GCGGGGCUGCGGAUGCAGAUGCAGAUGCAAUUGUCGCAACAUGUCGCCGCUGACUGUUGCAGCUGCAGAUGCA___GCAGUAGCAAUUGCAACUGCA .....(((((...((((..(((((.((((((.(((((...........)).))).)))))).)))))..))))...))))).((....))....... (-31.74 = -32.82 + 1.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:24 2006