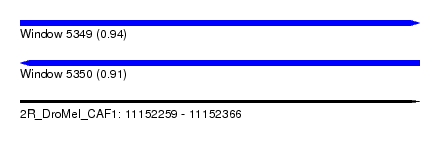
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,152,259 – 11,152,366 |
| Length | 107 |
| Max. P | 0.941506 |

| Location | 11,152,259 – 11,152,366 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.70 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -11.43 |
| Energy contribution | -11.07 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
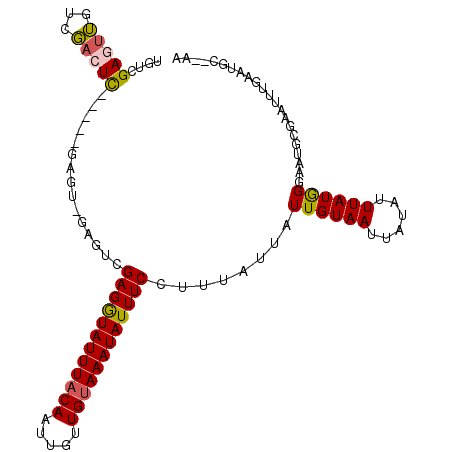
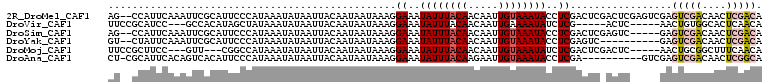
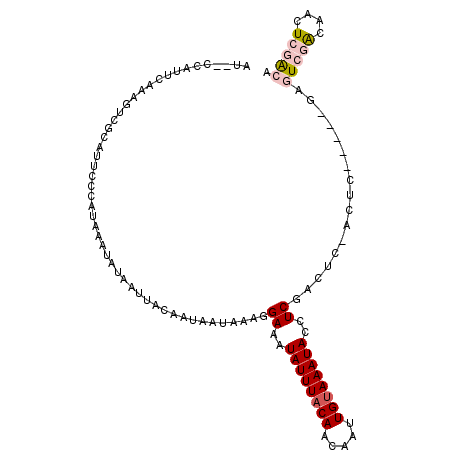
>2R_DroMel_CAF1 11152259 107 + 20766785 UGUCGAGUUGUCGACUCGACUCGAGUCGAGUCGAGGUAUUUACAAUUGUUGUAAAUAUUUCCUUUAUUAUUGUAAUUAUAUUUAUGGGAAUGCGAAUUUGAAUGG--CU ..(((((((.((((((((((....)))))))))).(((((((((.....)))))))))(((((....(((.......))).....)))))....)))))))....--.. ( -30.90) >DroVir_CAF1 98965 96 + 1 UGUUGAGUGCCACAGUU-----GAGU-----CGAGAUAUUUUCAAUUGUUGUAAAUAUUUCCUUUAUUAUUGUAAUUAUAUUUAUAGCUAUGUGGC---GGAUGCGGAA .((....((((((((((-----(((.-----.........)))))))(((((((((((.....((((....))))..)))))))))))...)))))---)...)).... ( -21.80) >DroSim_CAF1 65548 102 + 1 UGUCGAGUUGUCGACUC-----GACUCGAGUCGAGGUAUUUACAAUUGUUGUAAAUAUUUCCUUUAUUAUUGUAAUUAUAUUUAUGGGAAUGCGAAUUUGAAUGG--CU .((((((((...)))))-----)))(((((((((((((((((((.....))))))))))))...((((.(..(((......)))..).)))).).))))))....--.. ( -25.90) >DroYak_CAF1 66689 97 + 1 UGUCGAGUUGUCGACUC----------GACUCGAGGUAUUUACAAUUGUUGUAAAUAUUUCCUUUAUUAUUGUAAUUAUAUUUAUGGGAAUGCGAAUUUGAAUAG--AC .((((((((...)))))----------)))..((((((((((((.....))))))))))))...................(((((..((((....))))..))))--). ( -23.10) >DroMoj_CAF1 111603 98 + 1 UGUUGAAAGCCGCAGUU-----GAGUCGAGUCGAGAUAUUUACAAUUGUUGUAAAUAUUUCCUUUAUUAUUGUAAUUAUAUUUAUGGCCG---AAC---GGAAGCGGAA .........((((..((-----(..(((.(((((((((((((((.....))))))))))))..........((((......)))))))))---).)---))..)))).. ( -25.20) >DroAna_CAF1 66069 98 + 1 UGCCGAGUUGUCGACUCGAC----------UCGAGGUAUUUACAAUUCUUGUAAAUAUUUCCUUUAUUAUUGUAAUUAUAUUUAUGGGAAUGUGACUGUGAAUGCG-AG ...(((((((......))))----------)))..((((((((((((((..(((((((.....((((....))))..)))))))..))))).....))))))))).-.. ( -28.80) >consensus UGUCGAGUUGUCGACUC_____GAGU_GAGUCGAGGUAUUUACAAUUGUUGUAAAUAUUUCCUUUAUUAUUGUAAUUAUAUUUAUGGGAAUGCGAAUUUGAAUGC__AA ....(((((...)))))...............((((((((((((.....))))))))))))........((((((......))))))...................... (-11.43 = -11.07 + -0.36)

| Location | 11,152,259 – 11,152,366 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.70 |
| Mean single sequence MFE | -16.76 |
| Consensus MFE | -6.93 |
| Energy contribution | -7.73 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11152259 107 - 20766785 AG--CCAUUCAAAUUCGCAUUCCCAUAAAUAUAAUUACAAUAAUAAAGGAAAUAUUUACAACAAUUGUAAAUACCUCGACUCGACUCGAGUCGAGUCGACAACUCGACA ..--..........(((..............................((...((((((((.....))))))))))((((((((((....)))))))))).....))).. ( -23.50) >DroVir_CAF1 98965 96 - 1 UUCCGCAUCC---GCCACAUAGCUAUAAAUAUAAUUACAAUAAUAAAGGAAAUAUUUACAACAAUUGAAAAUAUCUCG-----ACUC-----AACUGUGGCACUCAACA ..........---(((((((((.(((....))).)))...........((.((((((.((.....)).)))))).)).-----....-----...))))))........ ( -11.90) >DroSim_CAF1 65548 102 - 1 AG--CCAUUCAAAUUCGCAUUCCCAUAAAUAUAAUUACAAUAAUAAAGGAAAUAUUUACAACAAUUGUAAAUACCUCGACUCGAGUC-----GAGUCGACAACUCGACA ..--...............((((........................)))).((((((((.....))))))))..(((((....)))-----))(((((....))))). ( -17.36) >DroYak_CAF1 66689 97 - 1 GU--CUAUUCAAAUUCGCAUUCCCAUAAAUAUAAUUACAAUAAUAAAGGAAAUAUUUACAACAAUUGUAAAUACCUCGAGUC----------GAGUCGACAACUCGACA ..--..........(((..((((........................)))).((((((((.....))))))))...)))(((----------((((.....))))))). ( -16.96) >DroMoj_CAF1 111603 98 - 1 UUCCGCUUCC---GUU---CGGCCAUAAAUAUAAUUACAAUAAUAAAGGAAAUAUUUACAACAAUUGUAAAUAUCUCGACUCGACUC-----AACUGCGGCUUUCAACA ..((((..((---...---.))..........................((.(((((((((.....))))))))).))..........-----....))))......... ( -14.40) >DroAna_CAF1 66069 98 - 1 CU-CGCAUUCACAGUCACAUUCCCAUAAAUAUAAUUACAAUAAUAAAGGAAAUAUUUACAAGAAUUGUAAAUACCUCGA----------GUCGAGUCGACAACUCGGCA .(-((.((((.........((((........................)))).((((((((.....))))))))....))----------)))))(((((....))))). ( -16.46) >consensus AU__CCAUUCAAAGUCGCAUUCCCAUAAAUAUAAUUACAAUAAUAAAGGAAAUAUUUACAACAAUUGUAAAUACCUCGACUC_ACUC_____GAGUCGACAACUCGACA ................................................((..((((((((.....))))))))..)).................(((((....))))). ( -6.93 = -7.73 + 0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:18 2006