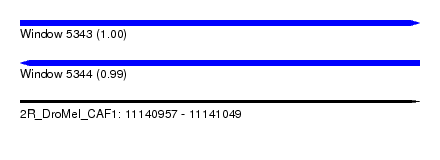
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,140,957 – 11,141,049 |
| Length | 92 |
| Max. P | 0.996608 |

| Location | 11,140,957 – 11,141,049 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 73.34 |
| Mean single sequence MFE | -34.51 |
| Consensus MFE | -22.21 |
| Energy contribution | -24.78 |
| Covariance contribution | 2.58 |
| Combinations/Pair | 1.34 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11140957 92 + 20766785 GCUGGGAAAAGUUGGUA-----AAAGUGGGGGGGCAAA-AUGGUGAAUGGCUGGCUAGCUGU-------AUAGCUGGCCAGUCAAGUGGCCAACUUUGUUGUGGC ((((.(((((((((((.-----...((......))...-........(((((((((((((..-------..)))))))))))))....))))))))).)).)))) ( -37.10) >DroSec_CAF1 55531 85 + 1 GCUGGGAAAAUUUGG-A-----AAAGUGG--GGGCAAA-AGGC--CAUGGGCCGCCACCUUU-------AAGG--GGGCACAACAGGGGGCAACUUUUGUGUGGC (((......((((..-.-----.))))..--.)))...-.(((--(...))))(((.(((..-------..))--))))(((.((((((....)))))))))... ( -26.30) >DroSim_CAF1 51814 90 + 1 GCUGGGAAAAGUUGGUA-----AAAGUGG--GGGCAAA-AUGGUGAAUGGCUGGCUAGCUGU-------AUAGCUGGCCAGUCAAGUGGCCAACUUUGUUGUGGC ((((.(((((((((((.-----...((..--..))...-........(((((((((((((..-------..)))))))))))))....))))))))).)).)))) ( -36.90) >DroEre_CAF1 58496 90 + 1 GCUGGGAAAAGUUGGUA-----AACGUGG--GGGCAAA-GUGGUGGAUGGCUGGCUAGCUGU-------AUAGCUGGCCAGUCAAGUGGGCAACUUUGUUGUGGC ((..(......)..)).-----...((.(--.((((((-((.((...(((((((((((((..-------..))))))))))))).....)).)))))))).).)) ( -34.90) >DroYak_CAF1 55010 103 + 1 GCUGGGAAAAGUUGGGAAAGCAAAGUUGG--GGGCAAACGUGGUGGAUGACUGGCUAGCUGUAUGCUAUAUAGCUGGCCAGUUAAGUGGCCAACUUUGUUGUGGC ((((.((((((((((....((...(((..--.....)))...))...(((((((((((((((((...))))))))))))))))).....)))))))).)).)))) ( -40.40) >DroAna_CAF1 54418 84 + 1 GGGCAGAAGAGGCGGCAGG------GCGA--GUGGAGA-AUGGAGGAGGGC----UA-CUGG-------CCAGUUGGCCAGUCAAGUGGCCAACUUUGUUGUGGC ...........((((((((------((.(--((((...-...........)----))-)).)-------))(((((((((......))))))))))))))))... ( -31.44) >consensus GCUGGGAAAAGUUGGUA_____AAAGUGG__GGGCAAA_AUGGUGGAUGGCUGGCUAGCUGU_______AUAGCUGGCCAGUCAAGUGGCCAACUUUGUUGUGGC ((((.(((((((((((.........((......)).............(((((((((((((((.....))))))))))))))).....))))))))).)).)))) (-22.21 = -24.78 + 2.58)

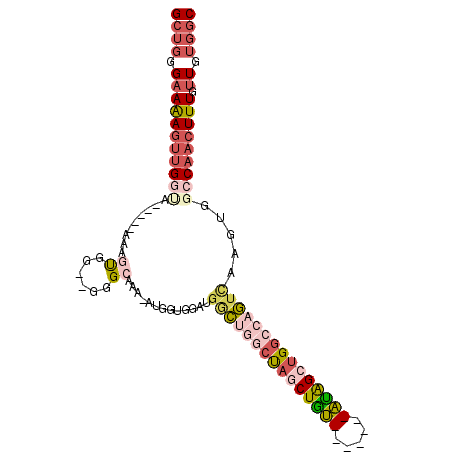
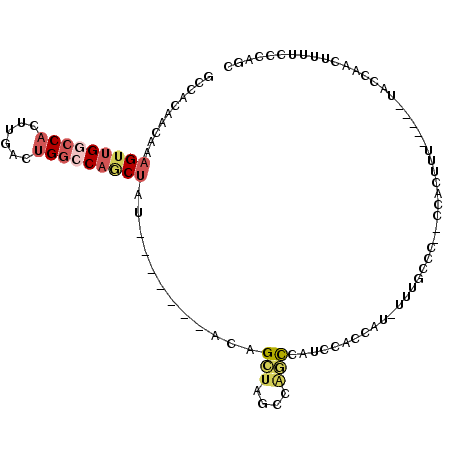
| Location | 11,140,957 – 11,141,049 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 73.34 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -9.18 |
| Energy contribution | -10.10 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11140957 92 - 20766785 GCCACAACAAAGUUGGCCACUUGACUGGCCAGCUAU-------ACAGCUAGCCAGCCAUUCACCAU-UUUGCCCCCCCACUUU-----UACCAACUUUUCCCAGC ........((((((((.....((.(((((.((((..-------..)))).))))).))........-..((......))....-----..))))))))....... ( -20.30) >DroSec_CAF1 55531 85 - 1 GCCACACAAAAGUUGCCCCCUGUUGUGCCC--CCUU-------AAAGGUGGCGGCCCAUG--GCCU-UUUGCCC--CCACUUU-----U-CCAAAUUUUCCCAGC (((..((((.((.......)).))))(((.--((..-------...)).))))))....(--((..-...))).--.......-----.-............... ( -16.10) >DroSim_CAF1 51814 90 - 1 GCCACAACAAAGUUGGCCACUUGACUGGCCAGCUAU-------ACAGCUAGCCAGCCAUUCACCAU-UUUGCCC--CCACUUU-----UACCAACUUUUCCCAGC ........((((((((.....((.(((((.((((..-------..)))).))))).))..((....-..))...--.......-----..))))))))....... ( -20.10) >DroEre_CAF1 58496 90 - 1 GCCACAACAAAGUUGCCCACUUGACUGGCCAGCUAU-------ACAGCUAGCCAGCCAUCCACCAC-UUUGCCC--CCACGUU-----UACCAACUUUUCCCAGC .......((((((........((.(((((.((((..-------..)))).))))).))......))-))))...--.......-----................. ( -18.34) >DroYak_CAF1 55010 103 - 1 GCCACAACAAAGUUGGCCACUUAACUGGCCAGCUAUAUAGCAUACAGCUAGCCAGUCAUCCACCACGUUUGCCC--CCAACUUUGCUUUCCCAACUUUUCCCAGC .......(((((((((.......((((((.((((.(((...))).)))).))))))((..(.....)..))...--))))))))).................... ( -24.20) >DroAna_CAF1 54418 84 - 1 GCCACAACAAAGUUGGCCACUUGACUGGCCAACUGG-------CCAG-UA----GCCCUCCUCCAU-UCUCCAC--UCGC------CCUGCCGCCUCUUCUGCCC ((........(((((((((......)))))))))((-------(..(-((----(..(........-.......--..).------.)))).)))......)).. ( -19.73) >consensus GCCACAACAAAGUUGGCCACUUGACUGGCCAGCUAU_______ACAGCUAGCCAGCCAUCCACCAU_UUUGCCC__CCACUUU_____UACCAACUUUUCCCAGC ..........(((((((((......)))))))))............(((....)))................................................. ( -9.18 = -10.10 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:12 2006