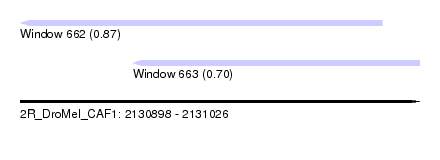
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,130,898 – 2,131,026 |
| Length | 128 |
| Max. P | 0.871804 |

| Location | 2,130,898 – 2,131,014 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.58 |
| Mean single sequence MFE | -46.10 |
| Consensus MFE | -26.34 |
| Energy contribution | -27.52 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
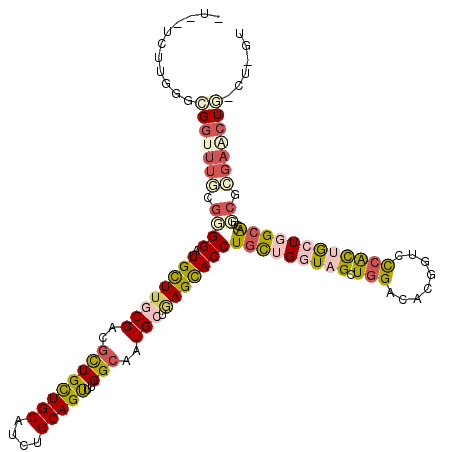
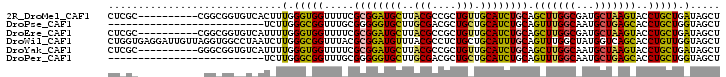
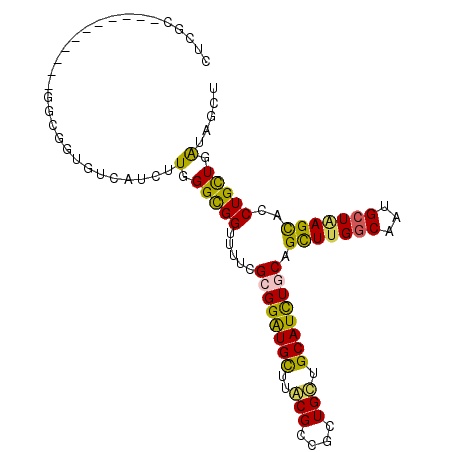
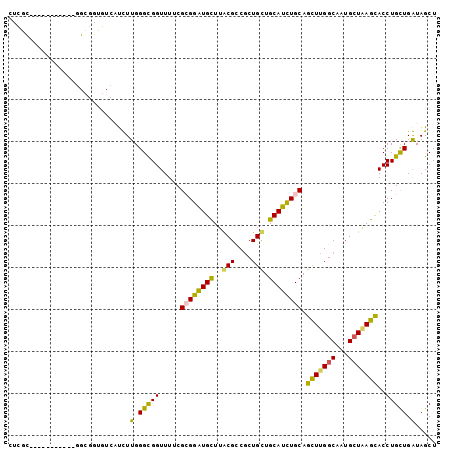
>2R_DroMel_CAF1 2130898 116 - 20766785 GUCACUUUGGGUGGUUUUCGCGGAUGCUUACGCCGCUGUUGCAUCUGCAGCUUGGCGAUGCUAAGUACCUGCUGAUAGCUGGACACCGUCUCACAACUCGCACUGCGCGAACUA-CU--- .........((((((((.(((((.(((...(((((..(((((....))))).))))).......((.((.((.....)).))))...............)))))))).))))))-))--- ( -40.60) >DroPse_CAF1 125818 115 - 1 ----UCUUGGGCGGUUUGCGGGGGUGCUUGCGACGCUGCUGCAUCUGCAGUUUGGCAAUGCUGAGCACCUGCUGGUAGCUGGACACGGUCCCACUGCUGGCAGUCCUCGAGCUGACC-GU ----.....(((((((((.(((((((((((((..((((((((....)))))..)))..))).))))))))((..((((..((((...))))..))))..))...)).))))))).))-.. ( -55.00) >DroGri_CAF1 227688 110 - 1 UUUGCUUUGGGCGGUUUUCUUGGAUGUUUGCGUCGCUGUUGCAUUUGCAGCUUGGCAAUGCUGAGCACCUGCUGAUAGCUGGACACGGUGCCGCUGCUUGAAC---------UA-AUUGU .(((.((..((((((..............((((.((((((((....)))))..))).)))).(.(((((((((.......)).)).))))))))))))..)).---------))-).... ( -33.50) >DroWil_CAF1 154322 119 - 1 CUAAUCUUGGGCGGUUUACGCGGAUGUUUACGCCUCUGCUGCAUUUGCAGUUUGGCUAUGGUCAGCACCUGUUGGUAGCUCGAAACGGUUUCAUUGCUGGCACUGCGUGAACUG-AUGCU ...........((((((((((((........(((...(((((....)))))..)))....(((((((((....))).....(((.....)))...)))))).))))))))))))-..... ( -44.40) >DroAna_CAF1 114318 113 - 1 ACCGUUUUGGGUGGCUUGCGGGGGUGCUUGCGGCGUUGCUGCAUCUGCAGUUUGGCGAUGCUGAGCACCUGUUGGUAGCUGGACACAGUUUCGCAGCUGGCGCUGCGCGAACU------- ...((.((.(((.(((.((..(((((((((((.(((((((((....)))))..)))).))).)))))))))).))).))).)).))(((((((((((....)))))).)))))------- ( -48.10) >DroPer_CAF1 124852 115 - 1 ----UCUUGGGCGGUUUGCGGGGGUGCUUGCGACGCUGCUGCAUCUGCAGUUUGGCAAUGCUGAGCACCUGCUGGUAGCUGGACACGGUCCCACUGCUGGCAGUCCUCGAGCUGACC-GU ----.....(((((((((.(((((((((((((..((((((((....)))))..)))..))).))))))))((..((((..((((...))))..))))..))...)).))))))).))-.. ( -55.00) >consensus _U__UCUUGGGCGGUUUGCGGGGAUGCUUGCGACGCUGCUGCAUCUGCAGUUUGGCAAUGCUGAGCACCUGCUGGUAGCUGGACACGGUCCCACUGCUGGCACUGCGCGAACUG_CU_GU ...........(((((((.((((.((((((((..((((((((....)))))..)))..))).)))))))(((((((((.(((........))))))))))))..)).)))))))...... (-26.34 = -27.52 + 1.17)

| Location | 2,130,934 – 2,131,026 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 75.25 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -22.32 |
| Energy contribution | -21.02 |
| Covariance contribution | -1.30 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.702115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2130934 92 - 20766785 CUCGC----------CGGCGGUGUCACUUUGGGUGGUUUUCGCGGAUGCUUACGCCGCUGUUGCAUCUGCAGCUUGGCGAUGCUAAGUACCUGCUGAUAGCU .((((----------((((((((((((.....)))......((....))..))))))))(((((....)))))..))))).(((((((....)))..)))). ( -33.80) >DroPse_CAF1 125857 76 - 1 --------------------------UCUUGGGCGGUUUGCGGGGGUGCUUGCGACGCUGCUGCAUCUGCAGUUUGGCAAUGCUGAGCACCUGCUGGUAGCU --------------------------.....(((..(..((..(((((((((((..((((((((....)))))..)))..))).))))))))))..)..))) ( -29.40) >DroEre_CAF1 108429 92 - 1 CUCGC----------CGGCGGUGUCAUUUUGGGUGGUUUUCGCGGAUGCUUACGCCGCUGUUGCAUCUGCAGCUUGGCGAUGCUAAGUACCUGCUGAUAGCU .((((----------((((((((((((((...(((.....))))))))...))))))))(((((....)))))..))))).(((((((....)))..)))). ( -33.40) >DroWil_CAF1 154361 102 - 1 CUGGUGAGGAUUGUUAGGUGGCCUAAUCUUGGGCGGUUUACGCGGAUGUUUACGCCUCUGCUGCAUUUGCAGUUUGGCUAUGGUCAGCACCUGUUGGUAGCU ...(((((.(((((((..(.(((((....))))).)..)).)).))).)))))(((...(((((....)))))..))).......((((((....))).))) ( -32.80) >DroYak_CAF1 110810 92 - 1 CUCGC----------GGGCGGUGUCAUUUUGGGUGGUUUUCGCGGAUGCUUACGCCGCUGUUGCAUCUGCAGCUUGGCAAUGCUAAGUACCUGCUGAUAGCU ...((----------...((((........((((.......((((((((..(((....))).)))))))).(((((((...)))))))))))))))...)). ( -29.70) >DroPer_CAF1 124891 76 - 1 --------------------------UCUUGGGCGGUUUGCGGGGGUGCUUGCGACGCUGCUGCAUCUGCAGUUUGGCAAUGCUGAGCACCUGCUGGUAGCU --------------------------.....(((..(..((..(((((((((((..((((((((....)))))..)))..))).))))))))))..)..))) ( -29.40) >consensus CUCGC___________GGCGGUGUCAUCUUGGGCGGUUUUCGCGGAUGCUUACGCCGCUGCUGCAUCUGCAGCUUGGCAAUGCUAAGCACCUGCUGAUAGCU .............................(.(((((.....((((((((..(((....))).)))))))).(((((((...)))))))..))))).)..... (-22.32 = -21.02 + -1.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:36 2006