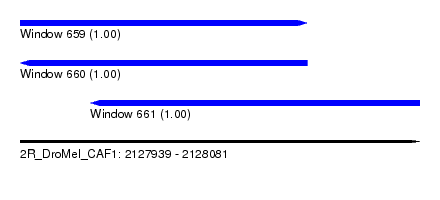
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,127,939 – 2,128,081 |
| Length | 142 |
| Max. P | 0.999770 |

| Location | 2,127,939 – 2,128,041 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 98.82 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.03 |
| SVM RNA-class probability | 0.999767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2127939 102 + 20766785 GCCACCACUGGCAGCCACAAACAAUAACAGUACGAACACUGCCACUCAGCCGCCGCCAUUGGCAGCUACGAACAAUACCAAUUCGAACACUGCCAACAAUUC ........((((((.............((((......))))......(((.((((....)))).))).((((.........))))....))))))....... ( -20.80) >DroSec_CAF1 106610 102 + 1 UCCACCACUGGCAGCCACAAACAAUAACAGUACGAACACUGCCACUCAGCCGCCGCCAUUGGCAGCUACGAACAAUACCAAUUCGAACACUGCCAACAAUUC ........((((((.............((((......))))......(((.((((....)))).))).((((.........))))....))))))....... ( -20.80) >DroSim_CAF1 110563 102 + 1 GCCACCACUGGCAGCCACAAACAAUAACAGUACGAACACUGCCACUCAGCCGCCGCCAUUGGCAGCUACGAACAAUACCAAUUCGAACACUGCCAACAAUUC ........((((((.............((((......))))......(((.((((....)))).))).((((.........))))....))))))....... ( -20.80) >DroEre_CAF1 105330 102 + 1 GCCACCACUGGCAGCCACAAACAAUAACAGUACGAACACUGCCACUCAGCCGCCGCCAUUGGCAGCUACAAACAAUACCAAUUCGAACACUGCCAACAAUUC ........((((((.............((((......))))......(((.((((....)))).)))......................))))))....... ( -20.20) >DroYak_CAF1 107666 102 + 1 GCCACCACUGGCAGCCACAAACAAUAACAGUACGAACACUGCCACUCAGCCGCCGCCAUUGGCAGCUACGAACAAUACCAAUUUGAACACUGCCAACAAUUC ........((((((...((((......((((......))))......(((.((((....)))).)))..............))))....))))))....... ( -20.80) >consensus GCCACCACUGGCAGCCACAAACAAUAACAGUACGAACACUGCCACUCAGCCGCCGCCAUUGGCAGCUACGAACAAUACCAAUUCGAACACUGCCAACAAUUC ........((((((.............((((......))))......(((.((((....)))).)))......................))))))....... (-20.20 = -20.20 + -0.00)

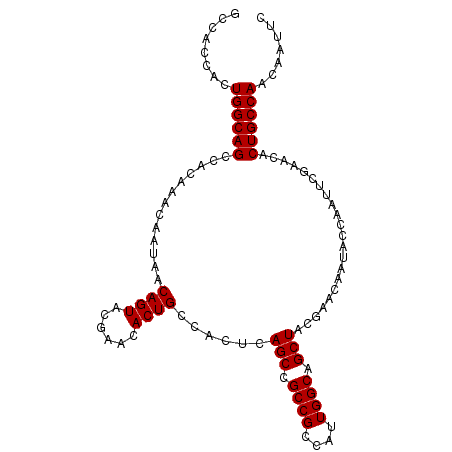
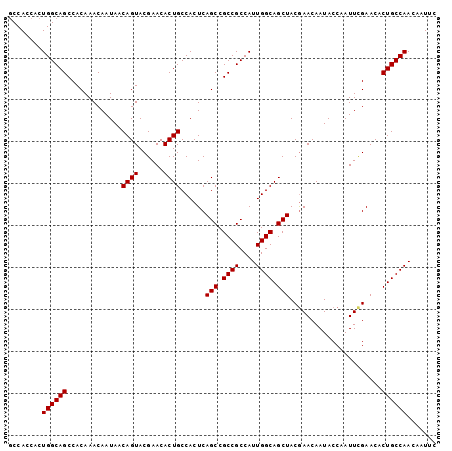
| Location | 2,127,939 – 2,128,041 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 98.82 |
| Mean single sequence MFE | -42.44 |
| Consensus MFE | -41.54 |
| Energy contribution | -41.62 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.68 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.999342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2127939 102 - 20766785 GAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUUAUUGUUUGUGGCUGCCAGUGGUGGC .....(((((((((...(((((.......)))))..)))))))))((((((.((..((((((((((....))))))))))....)).))))))......... ( -43.00) >DroSec_CAF1 106610 102 - 1 GAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUUAUUGUUUGUGGCUGCCAGUGGUGGA .....(((((((((...(((((.......)))))..)))))))))((((((.((..((((((((((....))))))))))....)).))))))......... ( -43.00) >DroSim_CAF1 110563 102 - 1 GAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUUAUUGUUUGUGGCUGCCAGUGGUGGC .....(((((((((...(((((.......)))))..)))))))))((((((.((..((((((((((....))))))))))....)).))))))......... ( -43.00) >DroEre_CAF1 105330 102 - 1 GAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUUGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUUAUUGUUUGUGGCUGCCAGUGGUGGC .....(((((((((...(((((.......)))))..)))))))))((((((.((..((((((((((....))))))))))....)).))))))......... ( -40.90) >DroYak_CAF1 107666 102 - 1 GAAUUGUUGGCAGUGUUCAAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUUAUUGUUUGUGGCUGCCAGUGGUGGC ((((...((.((((......)))).))..))))...((..(((.(((((((.((..((((((((((....))))))))))....)).))))))).)))..)) ( -42.30) >consensus GAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUUAUUGUUUGUGGCUGCCAGUGGUGGC .....(((((((((...(((((.......)))))..)))))))))((((((.((..((((((((((....))))))))))....)).))))))......... (-41.54 = -41.62 + 0.08)

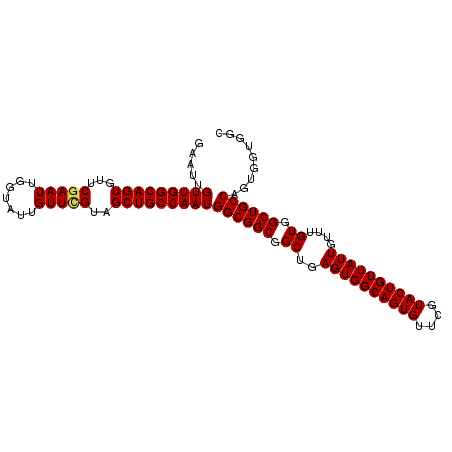
| Location | 2,127,964 – 2,128,081 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 98.97 |
| Mean single sequence MFE | -45.06 |
| Consensus MFE | -44.90 |
| Energy contribution | -44.98 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.17 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.04 |
| SVM RNA-class probability | 0.999770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2127964 117 - 20766785 AUUAUUCGAGGCAUUAUGGUUGAUACUGUUGCUGGCAGUCGAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUU .........((((.((((...((((((((..(((((.((((..(..((((((((...(((((.......)))))..))))))))..))))).))).))..)))))))))))).)))) ( -47.30) >DroSec_CAF1 106635 117 - 1 AUUAUUCGAGGCAUUAUGGUUGUUACUGUUGCUGGCAGUCGAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUU .........((((.(((((....((((((..(((((.((((..(..((((((((...(((((.......)))))..))))))))..))))).))).))..)))))).))))).)))) ( -46.50) >DroSim_CAF1 110588 117 - 1 AUUAUUCGAGGCAUUAUGGUUGUUACUGUUGCUGGCAGUCGAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUU .........((((.(((((....((((((..(((((.((((..(..((((((((...(((((.......)))))..))))))))..))))).))).))..)))))).))))).)))) ( -46.50) >DroEre_CAF1 105355 117 - 1 AUUAUUCGAGGCAUUAUGGUUGUUACUGUUGCUGGCAGUCGAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUUGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUU .........((((.(((((....((((((..(((((.((((..(..((((((((...(((((.......)))))..))))))))..))))).))).))..)))))).))))).)))) ( -44.40) >DroYak_CAF1 107691 117 - 1 AUUAUUCGAGGCAUUAUGGUUGUUACUGUUGCUGGCAGUCGAAUUGUUGGCAGUGUUCAAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUU .........((((.(((((....((((((..(((((.((((..(..((((((((...(.(((.......))).)..))))))))..))))).))).))..)))))).))))).)))) ( -40.60) >consensus AUUAUUCGAGGCAUUAUGGUUGUUACUGUUGCUGGCAGUCGAAUUGUUGGCAGUGUUCGAAUUGGUAUUGUUCGUAGCUGCCAAUGGCGGCGGCUGAGUGGCAGUGUUCGUACUGUU .........((((.(((((....((((((..(((((.((((..(..((((((((...(((((.......)))))..))))))))..))))).))).))..)))))).))))).)))) (-44.90 = -44.98 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:34 2006