| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,060,646 – 11,060,766 |
| Length | 120 |
| Max. P | 0.759899 |

| Location | 11,060,646 – 11,060,766 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.55 |
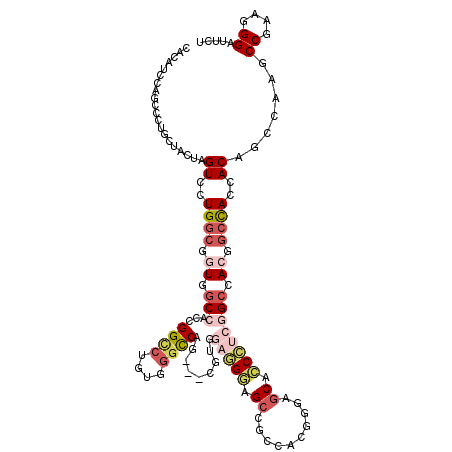
| Mean single sequence MFE | -48.82 |
| Consensus MFE | -21.11 |
| Energy contribution | -22.75 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
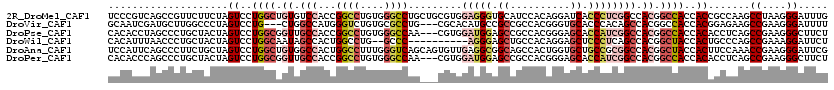
>2R_DroMel_CAF1 11060646 120 - 20766785 UCCCGUCAGCCGUUCUUCUAGUCCUGGCUGUGUCCACCGGCCUGUGGGCCUGCUGCGUGGAGGGUGCAUCCACAGGAUCACCCUCGGCCACGGCCACCACCGCCAAGCCUAAGGGAUUUG ((((..((((((.....(((((....)))).).....))).)))(((((.((.((.((((((((((.((((...)))))))))))(((....)))..))))).)).))))).)))).... ( -50.00) >DroVir_CAF1 8068 114 - 1 GCAAUCGAUGCUUGGCCCUAGUCCUG---CUGGCCAUGGGUCUGUGCGCCUG---CGCACAUGCCGCCGCCACGGGUGCACCCACAGCCACGGCCACCACGGAGAAGCCGAAGGGAUUUU (((.....)))(((((.....(((((---.(((((.(((((.((((((....---)))))).)))...((...(((....)))...)))).))))).)).)))...)))))......... ( -42.50) >DroPse_CAF1 7517 117 - 1 CACACCUAGCCCUGCUACUAGUCCUGGCGGUUGCCACCGGCCUGUGGGCCAA---CGUGGAUGGAGCCGCCACGGGAGCACCAUCGGCCACGGCCACCACACCUCAGCCGAAGGGCUUCU .......(((((((((.....(((((((((((.((((.((((....))))..---.))))....)))))))).))))))....(((((...((........))...)))))))))))... ( -49.90) >DroWil_CAF1 8199 108 - 1 CACAUUUAACCCUGCUACUAGUCCUGGCAAUAGCCACUGGCCUG--GCCC----------AGGGAGCUGCCACAGGAGCUCCCUCAGCCACGGCUACCACUGCCCAGCCGAAAGGAUUCU ...........(((....((((..((((....)))).(((((((--((..----------((((((((.(....).))))))))..)))).)))))..))))..)))((....))..... ( -41.00) >DroAna_CAF1 7632 120 - 1 UCCAUUCAGCCCUUCUGCUAGUCCUGGCUGUGGCCACUGGCCUUUGGGUCAGCAGUGUUGAGGCGGCAGCCACUGGUGCUGCCGCGGCCACGGCUACCACUUCCAAACCGAAGGGAUUCG .........((((((((..(((..(((((((((((.((((((....))))))..........(((((((((....).)))))))))))))))))))..)))..))....))))))..... ( -59.50) >DroPer_CAF1 7518 117 - 1 CACACCCAGCCCUGCUACUAGUCCUGGCGGUUGCCACCGGCCUGUGGGCCAA---CGUGGAUGGAGCCGCCACGGGAGCACCAUCGGCCACGGCCACCACACCUCAGCCGAAGGGCUUCU .......(((((((((.....(((((((((((.((((.((((....))))..---.))))....)))))))).))))))....(((((...((........))...)))))))))))... ( -50.00) >consensus CACAUCCAGCCCUGCUACUAGUCCUGGCGGUGGCCACCGGCCUGUGGGCCAG___CGUGGAGGGAGCCGCCACGGGAGCACCCUCGGCCACGGCCACCACAGCCAAGCCGAAGGGAUUCU ....................((..((((.((.(((...((((....)))).........(((((.((..........)).)))))))).)).))))..)).......((....))..... (-21.11 = -22.75 + 1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:50 2006