
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,056,225 – 11,056,358 |
| Length | 133 |
| Max. P | 0.882946 |

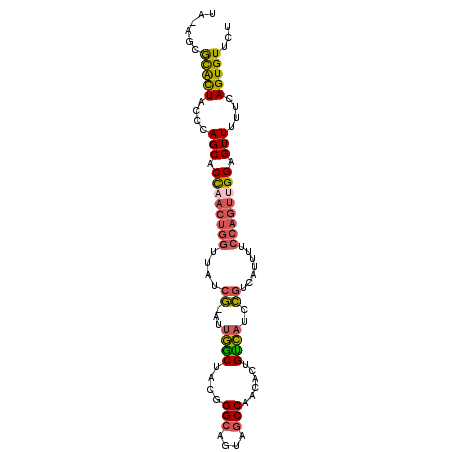
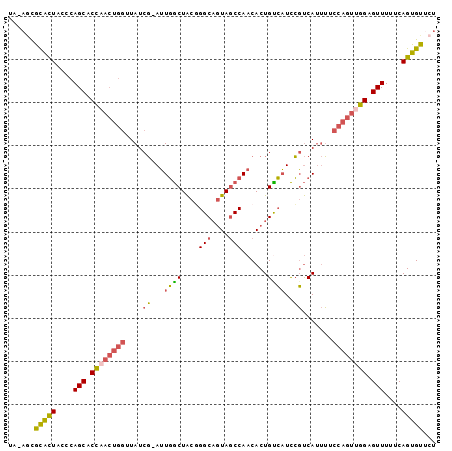
| Location | 11,056,225 – 11,056,322 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.58 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11056225 97 + 20766785 UA-AGCGCACUACCCAGCACCAACUGGUUAUCG-AUUGGCUACGGGCAGUAGCCAACACUGUCAUCCGUCAUUUUCCAGUUGGAGUUUUUCAGUGUUCU ..-((.(((((....(((.((((((((.....(-.((((((((.....)))))))))..((.(....).))....)))))))).)))....))))).)) ( -32.00) >DroVir_CAF1 3350 89 + 1 UGU-GUGUGUUUUGCAGCACUU------UAUCUAAGUUACUG-UGGCCUUAACCAACACUGUAUUUAUCCA--GUCCAGUUGGAGUUUUUCAGUGUUCU .(.-((.....(..(((.((((------.....))))..)))-..).....)))(((((((......((((--(.....)))))......))))))).. ( -18.90) >DroSec_CAF1 3045 97 + 1 UA-AGCGCACUACCCAGCACCAACUGGUUAUCG-AUUGGCUACGGGCAGUAGCCAACACUGUCAUCCGUCAUUUUCCAGUUGGAGUUUUUCAGUGUGCU ..-((((((((....(((.((((((((.....(-.((((((((.....)))))))))..((.(....).))....)))))))).)))....)))))))) ( -37.90) >DroSim_CAF1 2392 97 + 1 UA-AGCGCACUACCCAGCACCAACUGGUUAUCG-AUUGGCUACGGGCAGUAGCCAACACUGUCAUCCGUCAUUUUCCAGUAGGAGUUUUUCAGUGUUCU ..-((.(((((....(((.((.(((((.....(-.((((((((.....)))))))))..((.(....).))....))))).)).)))....))))).)) ( -27.90) >DroEre_CAF1 3052 97 + 1 UC-AACACACUUCCCAGCACCAACUGGUUAUCG-AUUGGCUACUGGCAGUAGCCAACACUGCCAUCCGUCAUUUUCCAGUUGGAGUUUUUCAGUGUUCU ..-...(((((....(((.((((((((....((-..((((...((((....)))).....))))..)).......)))))))).)))....)))))... ( -32.70) >DroYak_CAF1 3055 97 + 1 UC-AUCACACUUCCCAGCACCAACUGGUUAUCG-AUUGGCUACGGGCAGUAGCCAACACUGCCAUCCGUCAUUUUCCAGUUGGAGUUUUUCAGUGUUCU ..-...(((((....(((.((((((((....((-..((((....(((....)))......))))..)).......)))))))).)))....)))))... ( -32.20) >consensus UA_AGCGCACUACCCAGCACCAACUGGUUAUCG_AUUGGCUACGGGCAGUAGCCAACACUGUCAUCCGUCAUUUUCCAGUUGGAGUUUUUCAGUGUUCU ......(((((....(((.((((((((....((...((((....(((....)))......))))..)).......)))))))).)))....)))))... (-21.60 = -21.58 + -0.02)

| Location | 11,056,225 – 11,056,322 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -18.87 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11056225 97 - 20766785 AGAACACUGAAAAACUCCAACUGGAAAAUGACGGAUGACAGUGUUGGCUACUGCCCGUAGCCAAU-CGAUAACCAGUUGGUGCUGGGUAGUGCGCU-UA ((..(((((.......((((((((......((........))(((((((((.....)))))))))-......)))))))).......)))))..))-.. ( -32.14) >DroVir_CAF1 3350 89 - 1 AGAACACUGAAAAACUCCAACUGGAC--UGGAUAAAUACAGUGUUGGUUAAGGCCA-CAGUAACUUAGAUA------AAGUGCUGCAAAACACAC-ACA ..(((((((......((((.......--))))......)))))))(((....))).-(((((.(((.....------))))))))..........-... ( -18.60) >DroSec_CAF1 3045 97 - 1 AGCACACUGAAAAACUCCAACUGGAAAAUGACGGAUGACAGUGUUGGCUACUGCCCGUAGCCAAU-CGAUAACCAGUUGGUGCUGGGUAGUGCGCU-UA (((.(((((.......((((((((......((........))(((((((((.....)))))))))-......)))))))).......))))).)))-.. ( -35.84) >DroSim_CAF1 2392 97 - 1 AGAACACUGAAAAACUCCUACUGGAAAAUGACGGAUGACAGUGUUGGCUACUGCCCGUAGCCAAU-CGAUAACCAGUUGGUGCUGGGUAGUGCGCU-UA ..(((((((......(((....)))......(....).)))))))((((((((((((..((((((-.(.....).))))))..))))))))).)))-.. ( -31.80) >DroEre_CAF1 3052 97 - 1 AGAACACUGAAAAACUCCAACUGGAAAAUGACGGAUGGCAGUGUUGGCUACUGCCAGUAGCCAAU-CGAUAACCAGUUGGUGCUGGGAAGUGUGUU-GA ...(((((........((((((((....(((.((.((((((((.....))))))))....))..)-))....))))))))........)))))...-.. ( -33.29) >DroYak_CAF1 3055 97 - 1 AGAACACUGAAAAACUCCAACUGGAAAAUGACGGAUGGCAGUGUUGGCUACUGCCCGUAGCCAAU-CGAUAACCAGUUGGUGCUGGGAAGUGUGAU-GA ...(((((.....((((((.(((........))).))).)))(((((((((.....)))))))))-......(((((....)))))..)))))...-.. ( -31.90) >consensus AGAACACUGAAAAACUCCAACUGGAAAAUGACGGAUGACAGUGUUGGCUACUGCCCGUAGCCAAU_CGAUAACCAGUUGGUGCUGGGAAGUGCGCU_UA ..(((((((......(((....)))......(....).)))))))((((((.....))))))..........(((((....)))))............. (-18.87 = -19.82 + 0.95)

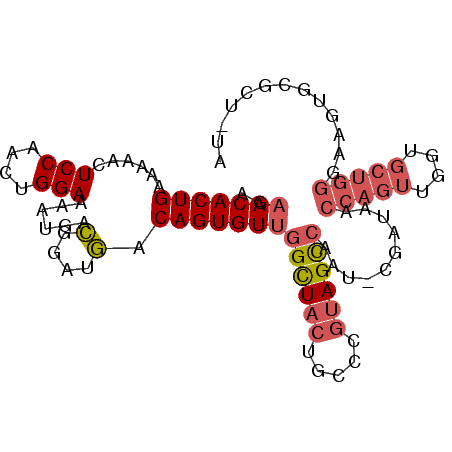
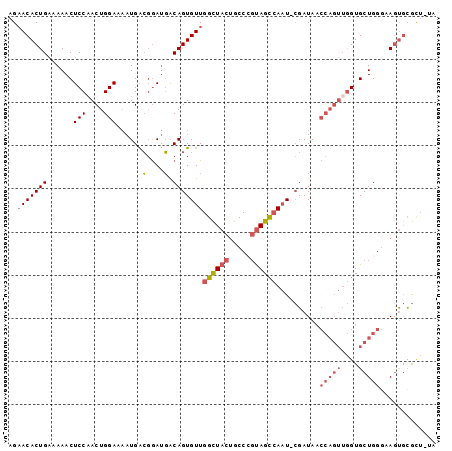
| Location | 11,056,246 – 11,056,358 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -32.19 |
| Consensus MFE | -13.31 |
| Energy contribution | -12.39 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11056246 112 + 20766785 ACUGGUUAUCGAUUGGCUACGGGCAGUAGCCAACACUGUCAUCCGUCAUUUUCCAGUUGGAGUUUUUCAGUGUUCUGCGACGGUUGCAGUUUUCUUGAUACGCAGUUGAAAA ((((((.(((((((((((((.....)))))))).((((.((.(((((...((((....)))).....(((....))).))))).))))))....))))))).))))...... ( -37.30) >DroPse_CAF1 3095 103 + 1 GUGUGCUAUCGAUUUCUUGCACUUGCUAUACAGCACUGC--------AAAUUCCAAUUGGAUUUUUUCAGUGU-CUUCGACUGGUGUAGUUUUUUUCGUGCGCAUUAGAAAA ((((((...(((....(((((((.........((((((.--------(((.(((....)))..))).))))))-........)))))))......))).))))))....... ( -26.83) >DroEre_CAF1 3073 112 + 1 ACUGGUUAUCGAUUGGCUACUGGCAGUAGCCAACACUGCCAUCCGUCAUUUUCCAGUUGGAGUUUUUCAGUGUUCUGCGACGGUUGCAGUUUUCUUGAUACGCAGUUGAGAA ((((((.(((((((((((((.....)))))))).(((((...(((((...((((....)))).....(((....))).)))))..)))))....))))))).))))...... ( -37.90) >DroYak_CAF1 3076 112 + 1 ACUGGUUAUCGAUUGGCUACGGGCAGUAGCCAACACUGCCAUCCGUCAUUUUCCAGUUGGAGUUUUUCAGUGUUCUGCGACGGUUGCAGUUUUCUUGAUACGCAGUUGAAAA ((((((.(((((((((((((.....)))))))).(((((...(((((...((((....)))).....(((....))).)))))..)))))....))))))).))))...... ( -38.40) >DroAna_CAF1 3268 108 + 1 AA-AGUUAUCGAUUUAUGGAAUGCGCUUAUCAACACUGGUGCCAGUCAU--UCCAGUUGGAGUUUUUUAGUGUUCUUCGACGG-UGCAGUUUUCUUGGUGCGCAGCAGAAAA ..-.(((.........(((((((.((((((((....)))))..))))))--))))(((((((..(......)..))))))).(-((((.(......).))))))))...... ( -26.40) >DroPer_CAF1 3099 103 + 1 GUGUGCUAUCGAUUUCUUGCAGUUGCUAGACAGCACUGC--------AAAUUCCAAUUGGAUUUUUUCAGUGU-CUUCGACUGGUGUAGUUUUUUUCGUGCGCAUUAGAAAA ((((((...(((....(((((((.(((....))))))))--------))((.(((((((((....))))))((-(...)))))).))........))).))))))....... ( -26.30) >consensus ACUGGUUAUCGAUUGGCUACAGGCACUAGCCAACACUGCCAUCCGUCAUUUUCCAGUUGGAGUUUUUCAGUGUUCUGCGACGGUUGCAGUUUUCUUGAUACGCAGUAGAAAA ........(((..........(((....)))(((((((.............(((....)))......)))))))...)))(((((((.((.........))))))))).... (-13.31 = -12.39 + -0.91)

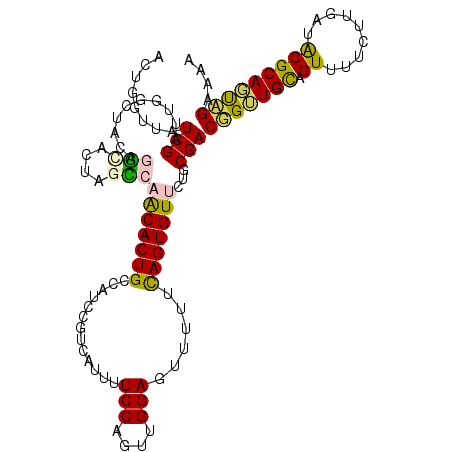
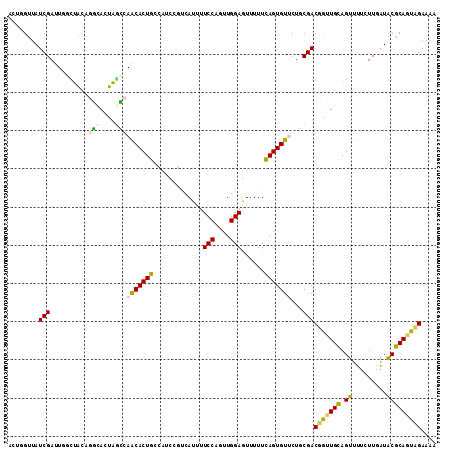
| Location | 11,056,246 – 11,056,358 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -7.61 |
| Energy contribution | -8.86 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.26 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11056246 112 - 20766785 UUUUCAACUGCGUAUCAAGAAAACUGCAACCGUCGCAGAACACUGAAAAACUCCAACUGGAAAAUGACGGAUGACAGUGUUGGCUACUGCCCGUAGCCAAUCGAUAACCAGU ......((((..((((......((((((.(((((((((....)))......(((....)))...)))))).)).))))(((((((((.....))))))))).))))..)))) ( -37.30) >DroPse_CAF1 3095 103 - 1 UUUUCUAAUGCGCACGAAAAAAACUACACCAGUCGAAG-ACACUGAAAAAAUCCAAUUGGAAUUU--------GCAGUGCUGUAUAGCAAGUGCAAGAAAUCGAUAGCACAC .(((((..(((((.(((...............))).((-.(((((...((((((....)).))))--------.))))))).........))))))))))............ ( -19.76) >DroEre_CAF1 3073 112 - 1 UUCUCAACUGCGUAUCAAGAAAACUGCAACCGUCGCAGAACACUGAAAAACUCCAACUGGAAAAUGACGGAUGGCAGUGUUGGCUACUGCCAGUAGCCAAUCGAUAACCAGU ......((((..((((......(((((..(((((((((....)))......(((....)))...))))))...)))))(((((((((.....))))))))).))))..)))) ( -39.20) >DroYak_CAF1 3076 112 - 1 UUUUCAACUGCGUAUCAAGAAAACUGCAACCGUCGCAGAACACUGAAAAACUCCAACUGGAAAAUGACGGAUGGCAGUGUUGGCUACUGCCCGUAGCCAAUCGAUAACCAGU ......((((..((((......(((((..(((((((((....)))......(((....)))...))))))...)))))(((((((((.....))))))))).))))..)))) ( -38.90) >DroAna_CAF1 3268 108 - 1 UUUUCUGCUGCGCACCAAGAAAACUGCA-CCGUCGAAGAACACUAAAAAACUCCAACUGGA--AUGACUGGCACCAGUGUUGAUAAGCGCAUUCCAUAAAUCGAUAACU-UU (((((.((.(.(((..........))).-).)).)))))..................((((--(((((((....))))(((....))).))))))).............-.. ( -19.80) >DroPer_CAF1 3099 103 - 1 UUUUCUAAUGCGCACGAAAAAAACUACACCAGUCGAAG-ACACUGAAAAAAUCCAAUUGGAAUUU--------GCAGUGCUGUCUAGCAACUGCAAGAAAUCGAUAGCACAC ........(((...(((............((((.....-..))))......(((....))).(((--------((((((((....))).))))))))...)))...)))... ( -22.20) >consensus UUUUCAACUGCGCACCAAGAAAACUGCAACCGUCGAAGAACACUGAAAAACUCCAACUGGAAAAUGACGGAUGGCAGUGUUGGCUACCGCCUGCAACAAAUCGAUAACCAGU ...............................((((...(((((((......(((....))).............)))))))(((....)))..........))))....... ( -7.61 = -8.86 + 1.25)

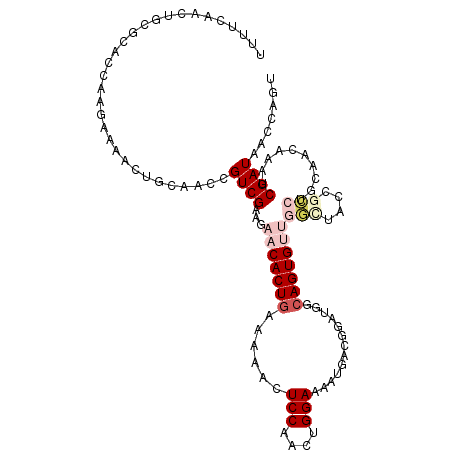
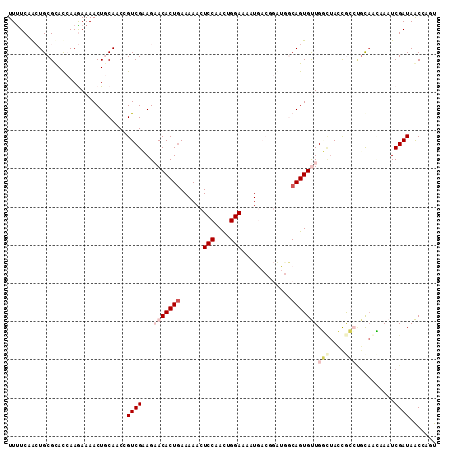
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:47 2006