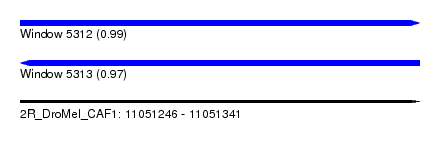
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,051,246 – 11,051,341 |
| Length | 95 |
| Max. P | 0.989809 |

| Location | 11,051,246 – 11,051,341 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 72.62 |
| Mean single sequence MFE | -17.55 |
| Consensus MFE | -10.85 |
| Energy contribution | -10.77 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
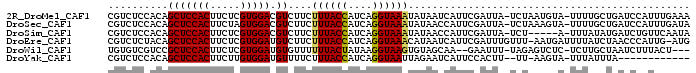
>2R_DroMel_CAF1 11051246 95 + 20766785 CGUCUCCACAGCUCCACUUCUCGUGGACGUCUUCUUUACCAUCAGGUAAAUAUAAUCAUUCGAUUA-UCUAAUGUA-UUUUGCUGAUCCAUUUGAAA ........((((.((((.....))))((((....((((((....)))))).((((((....)))))-)...)))).-....))))............ ( -18.50) >DroSec_CAF1 1185 95 + 1 CGUCUCCACAGCUCCACUUCUAGUGGACGUCUUCUUUACCAUCAGGUAAAUAUAACCAUUCGAUUA-UCUAAAGUA-UUUUGCUGAUCCAUUUGAUA ........(((((((((.....))))).((..(.((((((....)))))).)..))..........-.........-....))))............ ( -16.90) >DroSim_CAF1 1185 90 + 1 CGUCUCCACAGCUCCACUUCUCGUGGACGUCUUCUUUACCAUCAGGUAAAUAUAACCAUUCGAUUA-UCU-----A-UUUAUAUGAUCUGUUCAAUA ..........(((((((.....))))).))....((((((....))))))...........(((((-(..-----.-.....))))))......... ( -14.00) >DroEre_CAF1 1197 95 + 1 CGUCUCUACAGCUCCACUUCUCGUGGAUGUCUUCUUUACCAUCAGGUAAACAUAAUCAUUCGAUUUGUUU-AAUGAUUUUAUCUAACCCAUUG-AUG ((((......(((((((.....))))).))..........((((..((((((.((((....)))))))))-).))))...............)-))) ( -16.70) >DroWil_CAF1 1222 90 + 1 UGUGUCGUCCGCUCCACUUCUCGUGGAUGUGUUUUUUACUAUAAGGUAAGUGUAGCAA--GAAUUU-UAGAGUCUC-UCUUGCUAAUCUUUACU--- .(((..(..((((((((.....))))).)))..)..))).....((((((..((((((--((....-.........-))))))))...))))))--- ( -23.22) >DroYak_CAF1 1193 81 + 1 CGUCUCCACAGCUCCACUUCUUGUGGAUGUUUUCUUUACCAUCAGGUAAUUAGAAUCAUUCCACUU--UU-AAGUA-UUUAUUUA------------ ...............((((...(((((.(((((..(((((....)))))..)))))...)))))..--..-)))).-........------------ ( -16.00) >consensus CGUCUCCACAGCUCCACUUCUCGUGGACGUCUUCUUUACCAUCAGGUAAAUAUAACCAUUCGAUUA_UCU_AAGUA_UUUAGCUAAUCCAUUC_AUA ..........(((((((.....))))).))....((((((....))))))............................................... (-10.85 = -10.77 + -0.08)

| Location | 11,051,246 – 11,051,341 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 72.62 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -10.50 |
| Energy contribution | -10.67 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11051246 95 - 20766785 UUUCAAAUGGAUCAGCAAAA-UACAUUAGA-UAAUCGAAUGAUUAUAUUUACCUGAUGGUAAAGAAGACGUCCACGAGAAGUGGAGCUGUGGAGACG ............((((....-........(-(((((....)))))).((((((....)))))).......(((((.....))))))))).(....). ( -22.30) >DroSec_CAF1 1185 95 - 1 UAUCAAAUGGAUCAGCAAAA-UACUUUAGA-UAAUCGAAUGGUUAUAUUUACCUGAUGGUAAAGAAGACGUCCACUAGAAGUGGAGCUGUGGAGACG ............((((....-..((((..(-(((((....)))))).((((((....))))))))))...(((((.....))))))))).(....). ( -23.40) >DroSim_CAF1 1185 90 - 1 UAUUGAACAGAUCAUAUAAA-U-----AGA-UAAUCGAAUGGUUAUAUUUACCUGAUGGUAAAGAAGACGUCCACGAGAAGUGGAGCUGUGGAGACG ......((((.((.......-.-----..(-(((((....)))))).((((((....)))))))).....(((((.....))))).))))(....). ( -20.40) >DroEre_CAF1 1197 95 - 1 CAU-CAAUGGGUUAGAUAAAAUCAUU-AAACAAAUCGAAUGAUUAUGUUUACCUGAUGGUAAAGAAGACAUCCACGAGAAGUGGAGCUGUAGAGACG (((-((...(((.(((((.(((((((-..........))))))).)))))))))))))((.....((...(((((.....))))).))......)). ( -22.60) >DroWil_CAF1 1222 90 - 1 ---AGUAAAGAUUAGCAAGA-GAGACUCUA-AAAUUC--UUGCUACACUUACCUUAUAGUAAAAAACACAUCCACGAGAAGUGGAGCGGACGACACA ---.((.(((..((((((((-(........-...)))--))))))..))).((.....((.....))...(((((.....)))))..))))...... ( -17.30) >DroYak_CAF1 1193 81 - 1 ------------UAAAUAAA-UACUU-AA--AAGUGGAAUGAUUCUAAUUACCUGAUGGUAAAGAAAACAUCCACAAGAAGUGGAGCUGUGGAGACG ------------........-(((((-..--..(((((....((((..(((((....)))))))))....)))))...))))).......(....). ( -18.00) >consensus UAU_AAAUGGAUCAGAAAAA_UACUU_AGA_AAAUCGAAUGAUUAUAUUUACCUGAUGGUAAAGAAGACAUCCACGAGAAGUGGAGCUGUGGAGACG ................................................(((((....)))))........(((((.....)))))............ (-10.50 = -10.67 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:41 2006