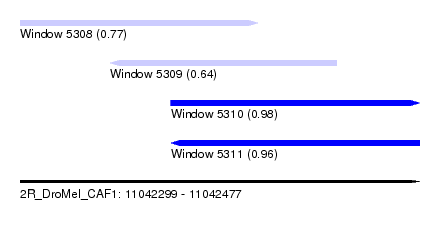
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,042,299 – 11,042,477 |
| Length | 178 |
| Max. P | 0.980654 |

| Location | 11,042,299 – 11,042,405 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 76.47 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -11.75 |
| Energy contribution | -13.80 |
| Covariance contribution | 2.05 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11042299 106 + 20766785 CGAAUGCGAAUGUCUUUGGUUGUUUUCUGUAUUAAAACGAAGGUUGAAAGG--CAUUAAGCAAA------CCGU--U--UGGCAUUACAAAGGCAUUACAGCGCGGCUAUAAGCCGCU .....((.((((((((((.((((((..(((.....(((....))).....)--))..)))))).------((..--.--.)).....))))))))))...))(((((.....))))). ( -35.40) >DroSec_CAF1 164920 106 + 1 CGAAUGCGAAUGUCUUUGGUUGUUUUCUGUAUUAAAACGAAGGUUGAAAGG--CAUUAAGCAAG------CCGU--U--UGGCAUUAAAAAGGCACUACAGUGUGGCUAUAGGCCGCU .....((.((((((..(((((((((..(((.....(((....))).....)--))..)))).))------))).--.--.)))))).....(((.(((.((.....)).)))))))). ( -30.60) >DroSim_CAF1 159168 106 + 1 CGAAUGCGAAUGUCUUUGGUUGUUUUCUGUAUUAAAACGAAGGUUGAAACG--CAUUAAGCAAG------CCGU--U--UGGCAUUAAAAAGGCAUUACAGUGUGGCUAUAAGCCGCU .....((.(((((((((..((((((..((..((..(((....)))..))..--))..))))))(------((..--.--.))).....)))))))))...))(((((.....))))). ( -31.70) >DroEre_CAF1 162131 102 + 1 CGAAGGCAAAUGUCUUUGGUUGUUUUCUGUAUUA-AACGAAGGUUGAGA-----UUUUAGCAAU------CCGU--U--UGGUAUUACAUAGGCAUUACAAUGUGGCUAUAAGCCACU (((((((....)))))))..(((((...((((((-((((..(((((...-----......))))------))))--)--)))))).....))))).......(((((.....))))). ( -28.20) >DroYak_CAF1 164110 105 + 1 CGAAUGCAAAUGUCUUUGGUUGUUUUCUGCAUUA-AACGAAGGUUGAAAAG--CAUUUAGCAAU------CAGU--U--UGGUAUUACAAAGGCAUUACAAUAUGGCUAUAAGCCACU ........((((((.(((((((((...(((....-(((....))).....)--))...))))))------)))(--(--((......))))))))))......((((.....)))).. ( -25.90) >DroPer_CAF1 150440 106 + 1 CGAAUGCAAAUGUCUUUGGUUGUUUUCUGUUUUUAGAUUGUGAAUCAAAAGAUCAUUAAUCAUAGAUACACUGUAUUUGUGAUAUCGUGACAUUAUUAUAGCAUGC------------ ...((((.(((((((((..(..(..((((....))))..)..)....))))).)))).(((((((((((...))))))))))).................))))..------------ ( -23.50) >consensus CGAAUGCAAAUGUCUUUGGUUGUUUUCUGUAUUAAAACGAAGGUUGAAAAG__CAUUAAGCAAA______CCGU__U__UGGCAUUACAAAGGCAUUACAGUGUGGCUAUAAGCCGCU ........((((((((((.((((((..........(((....)))............)))))).......(((......))).....)))))))))).....(((((.....))))). (-11.75 = -13.80 + 2.05)

| Location | 11,042,339 – 11,042,440 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.64 |
| Mean single sequence MFE | -19.65 |
| Consensus MFE | -13.87 |
| Energy contribution | -14.51 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11042339 101 - 20766785 AUUCACUGUAUGUUUCACAAUCAAUUUAAUUAUACAGCGGCUUAUAGCCGCGCUGUAAUGCCUUUGUAAUGCCAAACGGUUUGCUUAAUGCCUUUCAACCU ...........((((..((..(((....((((((..(((((.....)))))..))))))....)))...))..))))((((.((.....)).....)))). ( -20.30) >DroSec_CAF1 164960 101 - 1 AUUCGCUGUAUGUUUCAUAAUCAAUUUAAUUAAACAGCGGCCUAUAGCCACACUGUAGUGCCUUUUUAAUGCCAAACGGCUUGCUUAAUGCCUUUCAACCU ....(((((..(((..((.....))..)))...)))))(((((((((.....)))))).)))........(((....)))..((.....)).......... ( -20.70) >DroSim_CAF1 159208 101 - 1 AUUCGCUGUAUGUUUCAUAAUCAAUUUAAUUAAACAGCGGCUUAUAGCCACACUGUAAUGCCUUUUUAAUGCCAAACGGCUUGCUUAAUGCGUUUCAACCU ....(((((..(((..((.....))..)))...)))))(((((((((.....)))))).)))........(((....)))..((.....)).......... ( -19.20) >DroEre_CAF1 162170 98 - 1 AUCAGCUGUAUGUUACACAACCAAUUAAAUUAUACAGUGGCUUAUAGCCACAUUGUAAUGCCUAUGUAAUACCAAACGGAUUGCUAAAA---UCUCAACCU ....((((((((..................))))))))(((((((((.....)))))).)))...(((((.((....))))))).....---......... ( -17.17) >DroYak_CAF1 164149 101 - 1 GUCAGUUGUAUGUUACACAACCAAUUAAAUUAUACAGUGGCUUAUAGCCAUAUUGUAAUGCCUUUGUAAUACCAAACUGAUUGCUAAAUGCUUUUCAACCU (((((((...(((((((...........((((((..(((((.....)))))..)))))).....)))))))...))))))).((.....)).......... ( -20.89) >consensus AUUCGCUGUAUGUUUCACAAUCAAUUUAAUUAUACAGCGGCUUAUAGCCACACUGUAAUGCCUUUGUAAUGCCAAACGGAUUGCUUAAUGCCUUUCAACCU ....((((((((..................))))))))(((((((((.....)))))).)))...((((.(((....)))))))................. (-13.87 = -14.51 + 0.64)

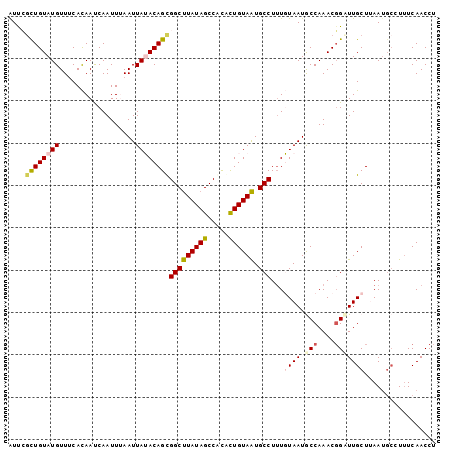
| Location | 11,042,366 – 11,042,477 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.09 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -14.40 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11042366 111 + 20766785 UGGCAUUACAAAGGCAUUACAGCGCGGCUAUAAGCCGCUGUAUAAUUAAAUUGAUUGUGAAACAUACAGUGAAUUAAAUAAUGCAGUAAAACGCACAUCAAAAUAUUGAGU ..((........(((.(((.(((...))).))))))(((((((.(((.((((.((((((.....)))))).)))).))).))))))).....))...((((....)))).. ( -25.60) >DroSec_CAF1 164987 111 + 1 UGGCAUUAAAAAGGCACUACAGUGUGGCUAUAGGCCGCUGUUUAAUUAAAUUGAUUAUGAAACAUACAGCGAAUUAAAUAAUGCAGCAAAACGCACAUCAAAAUAUUGAGU ..((((((....(((.(((.((.....)).))))))(((((.(((((.....)))))........)))))........)))))).((.....))...((((....)))).. ( -22.80) >DroSim_CAF1 159235 111 + 1 UGGCAUUAAAAAGGCAUUACAGUGUGGCUAUAAGCCGCUGUUUAAUUAAAUUGAUUAUGAAACAUACAGCGAAUUAAAUAAUGCAUUAAAACGCACAUCAAAAUAUUGAGU ..((((((...((.(((......))).)).(((..((((((.(((((.....)))))........))))))..)))..)))))).............((((....)))).. ( -19.30) >DroEre_CAF1 162194 91 + 1 UGGUAUUACAUAGGCAUUACAAUGUGGCUAUAAGCCACUGUAUAAUUUAAUUGGUUGUGUAACAUACAGCUGAUUAAAUAAUG--------------------UAAGCAGU .............((.(((((..(((((.....)))))......(((((((..(((((((...)))))))..)))))))..))--------------------)))))... ( -28.00) >DroYak_CAF1 164176 111 + 1 UGGUAUUACAAAGGCAUUACAAUAUGGCUAUAAGCCACUGUAUAAUUUAAUUGGUUGUGUAACAUACAACUGACUAAAUAAUGCAGUGAAACGCACUUGAAUGUGUGGAGU ...........((.(((......))).))......((((((((.(((((.(..(((((((...)))))))..).))))).))))))))...(((((......))))).... ( -32.30) >consensus UGGCAUUACAAAGGCAUUACAGUGUGGCUAUAAGCCGCUGUAUAAUUAAAUUGAUUGUGAAACAUACAGCGAAUUAAAUAAUGCAGUAAAACGCACAUCAAAAUAUUGAGU ..((((((.........((((..(((((.....)))))))))......((((.((((((.....)))))).))))...))))))........................... (-14.40 = -15.00 + 0.60)

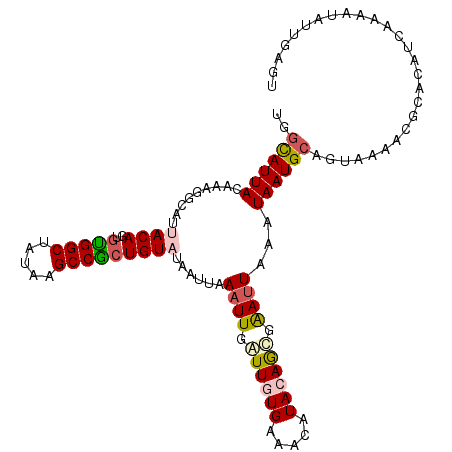
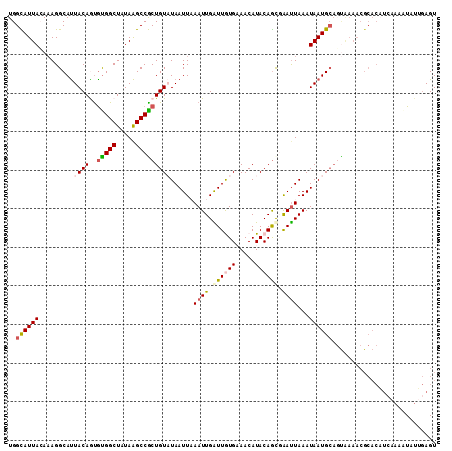
| Location | 11,042,366 – 11,042,477 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.09 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -15.85 |
| Energy contribution | -16.17 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11042366 111 - 20766785 ACUCAAUAUUUUGAUGUGCGUUUUACUGCAUUAUUUAAUUCACUGUAUGUUUCACAAUCAAUUUAAUUAUACAGCGGCUUAUAGCCGCGCUGUAAUGCCUUUGUAAUGCCA ..((((....))))((.(((((.....(((((((.........((((((..................))))))(((((.....)))))...)))))))......))))))) ( -24.47) >DroSec_CAF1 164987 111 - 1 ACUCAAUAUUUUGAUGUGCGUUUUGCUGCAUUAUUUAAUUCGCUGUAUGUUUCAUAAUCAAUUUAAUUAAACAGCGGCCUAUAGCCACACUGUAGUGCCUUUUUAAUGCCA ((((((....)))).))((.....)).((((((........(((((..(((..((.....))..)))...)))))(((((((((.....)))))).)))....)))))).. ( -23.30) >DroSim_CAF1 159235 111 - 1 ACUCAAUAUUUUGAUGUGCGUUUUAAUGCAUUAUUUAAUUCGCUGUAUGUUUCAUAAUCAAUUUAAUUAAACAGCGGCUUAUAGCCACACUGUAAUGCCUUUUUAAUGCCA ..((((....))))((.(((((..((.(((((((.......(((((..(((..((.....))..)))...)))))(((.....))).....)))))))..))..))))))) ( -22.70) >DroEre_CAF1 162194 91 - 1 ACUGCUUA--------------------CAUUAUUUAAUCAGCUGUAUGUUACACAACCAAUUAAAUUAUACAGUGGCUUAUAGCCACAUUGUAAUGCCUAUGUAAUACCA ...(((((--------------------((...........((((((((..................))))))))(((.....)))....))))).))............. ( -15.97) >DroYak_CAF1 164176 111 - 1 ACUCCACACAUUCAAGUGCGUUUCACUGCAUUAUUUAGUCAGUUGUAUGUUACACAACCAAUUAAAUUAUACAGUGGCUUAUAGCCAUAUUGUAAUGCCUUUGUAAUACCA .........(((((((.(((((.(((((.((.(((((((..(((((.......)))))..))))))).)).))))(((.....))).....).))))).)))).))).... ( -22.80) >consensus ACUCAAUAUUUUGAUGUGCGUUUUACUGCAUUAUUUAAUUCGCUGUAUGUUUCACAAUCAAUUUAAUUAUACAGCGGCUUAUAGCCACACUGUAAUGCCUUUGUAAUGCCA ...........................((((((........((((((((..................))))))))(((((((((.....)))))).)))....)))))).. (-15.85 = -16.17 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:39 2006