
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,015,158 – 11,015,261 |
| Length | 103 |
| Max. P | 0.610279 |

| Location | 11,015,158 – 11,015,261 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -13.78 |
| Energy contribution | -13.95 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11015158 103 + 20766785 AUACCUAUGCCCAGAAACUGAAUGCCAAACUGCAUUUCAUAGAUACAUCGAAGUUGGGGGCCAAGAAGCCCAGCGUCC-------UGGAUAUAAUGAACUGGGUGAAUGA ........((((((......(((((......)))))((((..((((...((.((((((..........)))))).)).-------..).))).)))).))))))...... ( -24.50) >DroSec_CAF1 127714 103 + 1 AUACCUAUGCCCAGAAACUGAAUGCCAAACUGCAGUUCAUAGAUACAGAGAAGUUGGGGGUCAAAAAGCCCAGCGUCC-------UAGACAUAAUGAACUGGGUGAAGGA ...(((.(((((((....(((((((......))).)))).......((.((.((((((..........)))))).)))-------)............))))))).))). ( -29.20) >DroSim_CAF1 130016 103 + 1 AUACCUAUGCCCAGAAACUGAAUGCCAAACUGCAGUUCAUAGACACAGAGAAGUUGGGGGUCAAGAAGCCCAGCGUCC-------UGGAUAUAAUGAACUGGGUGAAGGA ...(((.((((((.........(((......)))((((((..(..(((.((.((((((..(....)..)))))).)))-------))..)...)))))))))))).))). ( -33.00) >DroEre_CAF1 131010 103 + 1 ACACCUACGUCCAGAGACUGAAUGCCAAGCUGCAGUUUAUAGAUACAAUGGAGUUCGGGGUCAAGAAGGCCAGCGUCU-------UGGAUGUAAUGAAGUGGGCUAAGGA ...(((..(((((.((((((...((...))..)))))).....((((.(.(((..((.((((.....))))..)).))-------).).))))......)))))..))). ( -27.60) >DroYak_CAF1 133011 103 + 1 ACACCUAUGCCCAGAGACUGAAUGCCAAAUUGCAGUUUAUAAAUAUAACGGAGUUCGGGGUCAAGAAGGCCAGUGUCA-------UGGAUAUAAUGAAAUGGGCUAAGGA ...(((..(((((.(((((((((.....))).))))))...............((((.((((.....)))).(((((.-------..)))))..)))).)))))..))). ( -25.30) >DroPer_CAF1 127442 106 + 1 AUAGCUUCGCCCAGAAGGUGAAUGCAACCUUGGACUUUCGUUCCGUAAGGAAGGAUGG---CGAGAAGA-CAGAGUCCGUAUUUAAGGACAUAGUAAGAUGGGUGAAGGA ....(((((((((.(((((.......))))).(.((.((.((((....)))).)).))---).......-....((((........)))).........))))))))).. ( -36.40) >consensus AUACCUAUGCCCAGAAACUGAAUGCCAAACUGCAGUUCAUAGAUACAAAGAAGUUGGGGGUCAAGAAGCCCAGCGUCC_______UGGAUAUAAUGAACUGGGUGAAGGA ...(((.((((((.....(((...((.((((....................)))).))..)))...........((((........)))).........)))))).))). (-13.78 = -13.95 + 0.17)

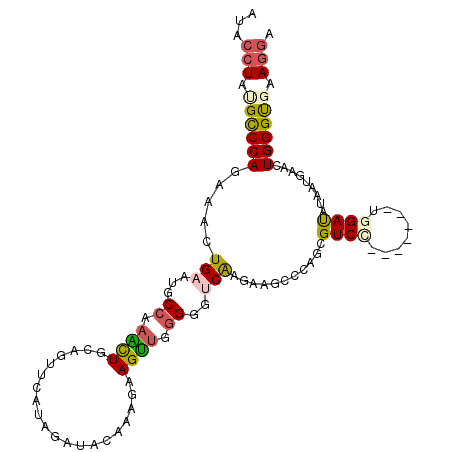
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:31 2006