
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 11,008,930 – 11,009,020 |
| Length | 90 |
| Max. P | 0.967104 |

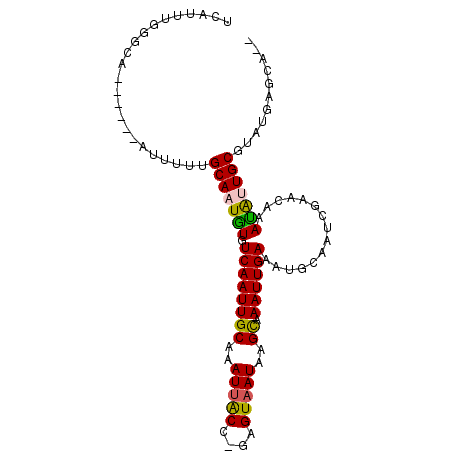
| Location | 11,008,930 – 11,009,020 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -14.17 |
| Energy contribution | -13.67 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
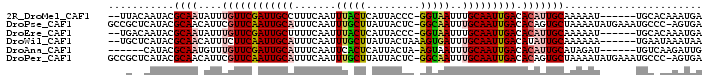
>2R_DroMel_CAF1 11008930 90 + 20766785 UCAUUUGUGCA------AUUUUUGCAAUGUGUCAAUUGCAAAUUACC-GGGUAAUGAGUAAAUUGAAAGGCAAUCGAACAAAUAUUGCGUAUUGUAA-- .......((((------((...((((((((.((.(((((.(((((((-(.....)).)).)))).....))))).))....)))))))).)))))).-- ( -19.30) >DroPse_CAF1 144344 97 + 1 UCACU-GGGCAUUUCAUAUUUUAGCACUGUGUCAAUUGCAAAUUGCC-GAGUAAUAAGCAAAUUGAAAUGCAAUUGAACGAAUGUUGCGUAUGAGCGGC .....-..((..(((((((..(((((...(((((((((((..((((.-.(....)..)))).......)))))))).)))..))))).)))))))..)) ( -21.50) >DroEre_CAF1 124695 90 + 1 UCAUUUGUGCA------AUUUUUGCAAUGUGUCAAUUGCAAAUUACC-GGGUAAUGAGUAAAUUGAAAAGCAAUCGAACAAAUAUUGCGUAUUGUCA-- ........(((------((...((((((((.((.(((((...(((((-(.....)).))))..(....)))))).))....)))))))).)))))..-- ( -17.60) >DroWil_CAF1 171790 91 + 1 UUAUUUAUUCA------UUUUUUGCAAUAUGUCAAUUGCAAAUCACUUUAGUAAUAAGCAAAUUGAAAUGCAAUUGAAGAAAUGUUGCGUAUGAGCA-- .......((((------(....((((((((.(((((((((..(((.(((.((.....))))).)))..)))))))))....)))))))).)))))..-- ( -24.50) >DroAna_CAF1 126217 86 + 1 CAAUCUUGACA------AUCUAUGCAAUGUGUCAAUUGCAAAUUACU-UAGUAAUGAGUGAAUUGAAAUGCAAUCGAACAAACAUUGCGUAUG------ ...........------...((((((((((.((.((((((..(((((-((....))))))).......)))))).))....))))))))))..------ ( -24.10) >DroPer_CAF1 121377 97 + 1 UCACU-GGGCAUUUCAUAUUUUAGCACUGUGUCAAUUGCAAAUUGCC-GAGUAAUAAGCAAAUUGAAAUGCAAUUGAACGAAUGUUGCGUAUGAGCGGC .....-..((..(((((((..(((((...(((((((((((..((((.-.(....)..)))).......)))))))).)))..))))).)))))))..)) ( -21.50) >consensus UCAUUUGGGCA______AUUUUUGCAAUGUGUCAAUUGCAAAUUACC_GAGUAAUAAGCAAAUUGAAAUGCAAUCGAACAAAUAUUGCGUAUGAGCA__ .......................(((((((.((((((((..(((((....)))))..)).))))))...............)))))))........... (-14.17 = -13.67 + -0.50)

| Location | 11,008,930 – 11,009,020 |
|---|---|
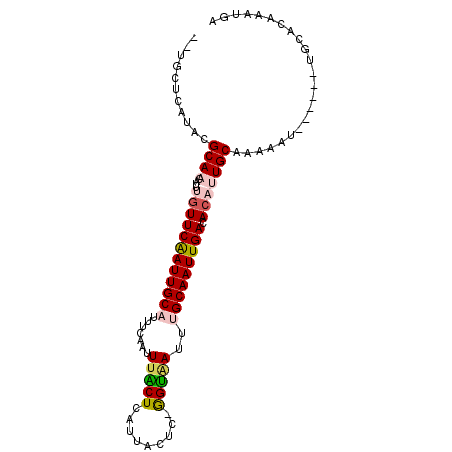
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -18.62 |
| Consensus MFE | -12.52 |
| Energy contribution | -12.80 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 11008930 90 - 20766785 --UUACAAUACGCAAUAUUUGUUCGAUUGCCUUUCAAUUUACUCAUUACCC-GGUAAUUUGCAAUUGACACAUUGCAAAAAU------UGCACAAAUGA --.........(((((.(((((((((((((.....((.(((((........-))))).))))))))))......))))).))------)))........ ( -17.60) >DroPse_CAF1 144344 97 - 1 GCCGCUCAUACGCAACAUUCGUUCAAUUGCAUUUCAAUUUGCUUAUUACUC-GGCAAUUUGCAAUUGACACAGUGCUAAAAUAUGAAAUGCCC-AGUGA ((...(((((.....((((.(((((((((((.......(((((........-)))))..))))))))).))))))......)))))...))..-..... ( -19.40) >DroEre_CAF1 124695 90 - 1 --UGACAAUACGCAAUAUUUGUUCGAUUGCUUUUCAAUUUACUCAUUACCC-GGUAAUUUGCAAUUGACACAUUGCAAAAAU------UGCACAAAUGA --.........(((((.(((((((((((((........(((((........-)))))...))))))))......))))).))------)))........ ( -18.00) >DroWil_CAF1 171790 91 - 1 --UGCUCAUACGCAACAUUUCUUCAAUUGCAUUUCAAUUUGCUUAUUACUAAAGUGAUUUGCAAUUGACAUAUUGCAAAAAA------UGAAUAAAUAA --(((......)))..((((.((((.(((((..((((((.((..(((((....)))))..)))))))).....)))))....------)))).)))).. ( -18.80) >DroAna_CAF1 126217 86 - 1 ------CAUACGCAAUGUUUGUUCGAUUGCAUUUCAAUUCACUCAUUACUA-AGUAAUUUGCAAUUGACACAUUGCAUAGAU------UGUCAAGAUUG ------.....(((((((....(((((((((....((((.(((........-)))))))))))))))).)))))))......------........... ( -18.50) >DroPer_CAF1 121377 97 - 1 GCCGCUCAUACGCAACAUUCGUUCAAUUGCAUUUCAAUUUGCUUAUUACUC-GGCAAUUUGCAAUUGACACAGUGCUAAAAUAUGAAAUGCCC-AGUGA ((...(((((.....((((.(((((((((((.......(((((........-)))))..))))))))).))))))......)))))...))..-..... ( -19.40) >consensus __UGCUCAUACGCAACAUUUGUUCAAUUGCAUUUCAAUUUACUCAUUACUC_GGUAAUUUGCAAUUGACACAUUGCAAAAAU______UGCACAAAUGA ...........((((....((((((((((((.......(((((.........)))))..))))))))).)))))))....................... (-12.52 = -12.80 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:29 2006