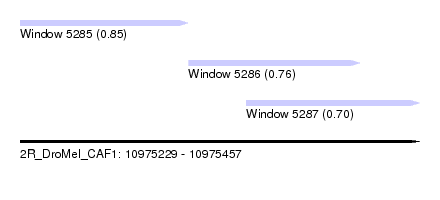
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,975,229 – 10,975,457 |
| Length | 228 |
| Max. P | 0.854696 |

| Location | 10,975,229 – 10,975,325 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 76.98 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -13.70 |
| Energy contribution | -14.98 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10975229 96 + 20766785 GCGCACAUAAUUAGCGCCACAGCUCUUGGCGCUGCACACUGGCAACUAGCAACUAGCAACCUGCAUCCGGCAACCUGCAACCAG-------CCAUCUGCAACA ..(((......((((((((.......)))))))).....((((.....((.....))....((((...(....).))))....)-------)))..))).... ( -26.60) >DroSec_CAF1 87604 81 + 1 GCGCACAUAAUUAGCGCCACAGCUCUUGGCGCUGCACACUGGCAACUAGCA--------CCUGCAUC-GGCA--CUGCA-CCAG-------CAU--UGC-ACA ..((.......((((((((.......)))))))).......)).....((.--------...))...-.(((--.(((.-...)-------)).--)))-... ( -23.34) >DroSim_CAF1 89887 96 + 1 GCGCACAUAAUUAGCGCCACAGCUCUUGGCGCUGCACACUGGCAACUAGCAACUAGCAACCUGCAUCCGGCAACCUGCAACCAG-------CCAUCUGCAACA ..(((......((((((((.......)))))))).....((((.....((.....))....((((...(....).))))....)-------)))..))).... ( -26.60) >DroEre_CAF1 89951 95 + 1 GCGCACAUAAUUAGCGCCACAACUCUU--------ACACUGGCAACUGGGAGCCUGCAUCCUGCAACCUGCAACCUGCAACCAGCAAACUGCAAUCUGCAGCG ((((.........)))).....(((((--------(....(....)))))))........(((((...((((...(((.....)))...))))...))))).. ( -21.90) >DroYak_CAF1 91510 96 + 1 GCGCACAUAAUUAGCGCCACAUCUCUUGGCGCUACACACUGGCAACUAGGAACCUGCAUCCUGCAACCUGCAAC-------CAGCAACCGGCAGUCUGCAACG ..((.......((((((((.......)))))))).......))...........((((..((((....(((...-------..)))....))))..))))... ( -29.74) >consensus GCGCACAUAAUUAGCGCCACAGCUCUUGGCGCUGCACACUGGCAACUAGCAACCAGCAACCUGCAUCCGGCAACCUGCAACCAG_______CAAUCUGCAACA ((((.......((((((((.......)))))))).......))..................(((.....))).........................)).... (-13.70 = -14.98 + 1.28)

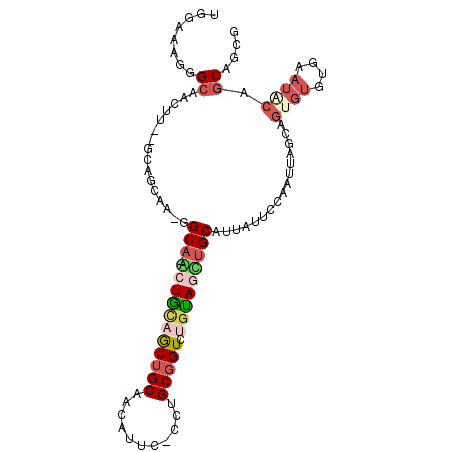
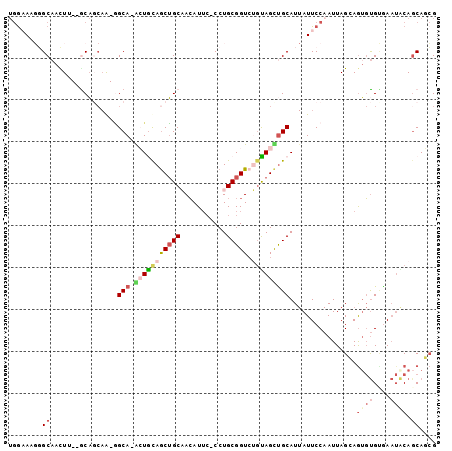
| Location | 10,975,325 – 10,975,423 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -10.05 |
| Energy contribution | -11.30 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10975325 98 + 20766785 UGGAAUGGGCAACUU--GCACCAA-GGCAGGCUGUAGCUGCAACAUUC-CCUGCGGUCUGUAGUUGCAUUAUUCCAAUUAGCAGUGUGCGAAUACAGCAGCG ((((((((((((((.--.......-.(((((((((((...........-.))))))))))))))))).))))))))....((.((((....)))).)).... ( -33.20) >DroSec_CAF1 87685 83 + 1 UGA-AU--GCCACUU--GCAC--A-AGCA-GCUGUAGCUGCUAAAU---CCUGCGGUC-GUAGCUGCAU-AUUCC-AUUAGCAGUUCG--A--UCAGCAGCG ...-.(--((.....--))).--.-((((-((....))))))....---.((((((((-(.((((((.(-(....-..))))))))))--)--)).)))).. ( -29.90) >DroSim_CAF1 89983 97 + 1 UGGAAUGGGCAAUUU--GCAGCAA-GGCA-GCUGUAGCUGCAACAUUC-CCUGCGGUCUGUAGCUGCAUUAUUCCAAUUAGCAGUGUGCGAAUACAUCAGCG ((((((((((.....--)).....-.(((-((((((((((((......-..))))).)))))))))).))))))))....((.(((((....)))))..)). ( -33.60) >DroEre_CAF1 90046 97 + 1 UGGAAAGGGCAACUA--GCAGCAG-GGCA-AGUACAGCUGCAACAUUC-CCUGCGGUGUGUAGUCGCAUUAUUCCAAUUAGGAGUGUGUGAAUACAGCACCG .((((.((....)).--(((((.(-....-....).)))))....)))-)...(((((((((.((((((.(((((.....))))))))))).)))).))))) ( -37.00) >DroYak_CAF1 91606 97 + 1 UGGAAAGGGCAAUUG--ACAGCAA-GGCA-AGUACAGCUGCAACAUUC-CCUGCGGUCUGUAGUUGCAUUAUUCCAAUUAGGAGUGUGUGAAUGCAGCACCG ......((((.(((.--(((....-.(((-(.((((((((((......-..))))).))))).))))..((((((.....))))))))).)))...)).)). ( -33.00) >DroAna_CAF1 90328 99 + 1 --GUAACUGCAACUGCAACUGCAAGUGCA-ACUGCAACGGCCACAUUCCCCUGCGGUCGUCAGCUGCAUUAUUCCAAUUAGGAAAGUGUAAAUGCAGCAAGG --(((..((((..(((....)))..))))-..))).((((((.((......)).))))))..((((((((.((((.....))))......)))))))).... ( -33.20) >consensus UGGAAAGGGCAACUU__GCAGCAA_GGCA_ACUGCAGCUGCAACAUUC_CCUGCGGUCUGUAGCUGCAUUAUUCCAAUUAGCAGUGUGUGAAUACAGCAGCG ........((................(((.(((((((((((...........))))).)))))))))................((((....)))).)).... (-10.05 = -11.30 + 1.25)
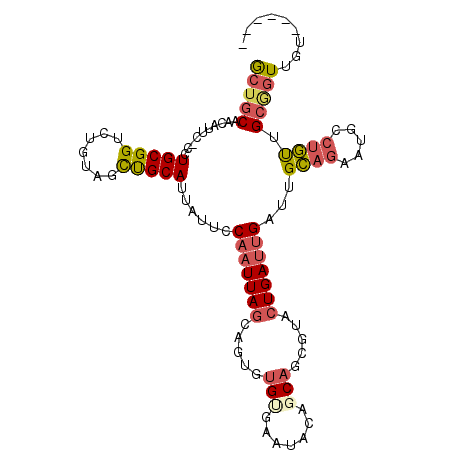
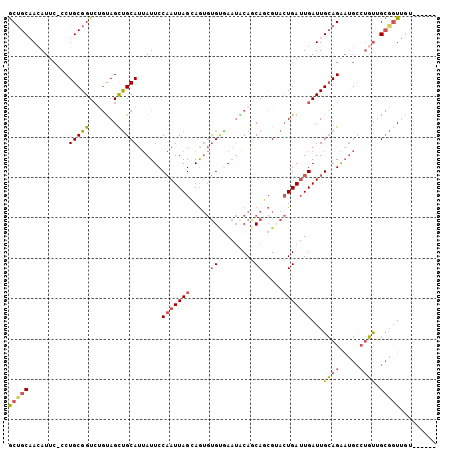
| Location | 10,975,358 – 10,975,457 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.09 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -13.90 |
| Energy contribution | -14.60 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10975358 99 + 20766785 GCUGCAACAUUC-CCUGCGGUCUGUAGUUGCAUUAUUCCAAUUAGCAGUGUGCGAAUACAGCAGCGUACUGAUUGAUUGCAGAAUGCCUGUUGCGGUUGU------ (((((((((...-.((((.....))))..(((((....(((((((((((((((..........)))))))).)))))))...))))).)))))))))...------ ( -32.20) >DroSec_CAF1 87712 87 + 1 GCUGCUAAAU---CCUGCGGUC-GUAGCUGCAU-AUUCC-AUUAGCAGUUCG--A--UCAGCAGCGUA-UGA-UGAUUGCAGAACGUCUGUUGCAGUGU------- (((((...((---.((((((((-(.((((((.(-(....-..))))))))))--)--)).)))).)).-...-.....((((.....)))).)))))..------- ( -29.20) >DroSim_CAF1 90015 99 + 1 GCUGCAACAUUC-CCUGCGGUCUGUAGCUGCAUUAUUCCAAUUAGCAGUGUGCGAAUACAUCAGCGUACUGAUUGAUUGCAGAAUGCCUGUGGCGGUUGC------ ((..((.(((((-..((((((.....))))))......(((((((((((((((..........)))))))).)))))))..)))))..))..))......------ ( -31.30) >DroEre_CAF1 90078 99 + 1 GCUGCAACAUUC-CCUGCGGUGUGUAGUCGCAUUAUUCCAAUUAGGAGUGUGUGAAUACAGCACCGAACUGAUUGAUUGCAGGAUGUCUGUUGCGGUUGU------ (((((((((..(-(((((((((((((.((((((.(((((.....))))))))))).)))).))))...(.....)...)))))..)..)))))))))...------ ( -41.20) >DroYak_CAF1 91638 99 + 1 GCUGCAACAUUC-CCUGCGGUCUGUAGUUGCAUUAUUCCAAUUAGGAGUGUGUGAAUGCAGCACCGAACUGAUUGAUUGCAGAAUGUGAGUUGCGGUUGU------ ((((((((....-....(((((((((.(..(((.(((((.....))))))))..).))))).))))..(((........))).......))))))))...------ ( -35.00) >DroAna_CAF1 90361 106 + 1 ACGGCCACAUUCCCCUGCGGUCGUCAGCUGCAUUAUUCCAAUUAGGAAAGUGUAAAUGCAGCAAGGCACUGAUUGAUUGGAGAUUGCCUUCAGGACUGCCUCAUCC ((((((.((......)).))))))..((((((((.((((.....))))......)))))))).(((((....((((..((......)).))))...)))))..... ( -32.40) >consensus GCUGCAACAUUC_CCUGCGGUCUGUAGCUGCAUUAUUCCAAUUAGCAGUGUGUGAAUACAGCAGCGUACUGAUUGAUUGCAGAAUGCCUGUUGCGGUUGU______ (((((..........(((((.......)))))......(((((((.....(((.......))).....)))))))...((((.....)))).)))))......... (-13.90 = -14.60 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:14 2006