| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,974,047 – 10,974,152 |
| Length | 105 |
| Max. P | 0.699051 |

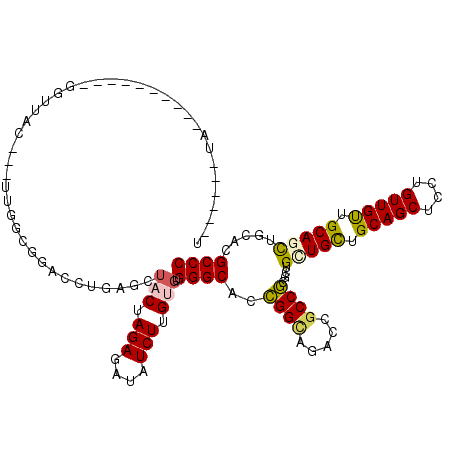
| Location | 10,974,047 – 10,974,152 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.15 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -25.20 |
| Energy contribution | -24.95 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
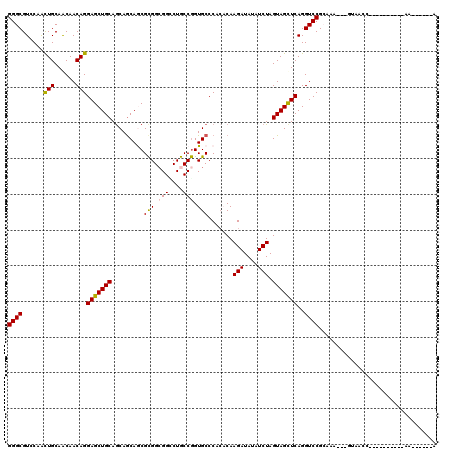
>2R_DroMel_CAF1 10974047 105 + 20766785 GGGCGUACAACUGCAGCAACAGGAGCUGCAGCAGCAGCGCGGCGGACUGCCGGUGCCCAGCCAAGAUAUAUCUAGUAGCUCAGGUCCGACAA---GUCGCCA------AGUUGC------ ..........(((((((.......)))))))..(((((..(((((..((.(((.(((.(((..(((....)))....)))..)))))).)).---.))))).------.)))))------ ( -38.00) >DroVir_CAF1 111960 108 + 1 GGGCGUCCAAUUGCAGCAACAGGAGCUGCAGCAGCAGCGCGGCGGUCUGCCGGUGCCCACACAAGAUAUCUCUAGUAGCUCAGGUCCGCAAAAAAAAAACCA------AAAAA------A (((((......))).((.((..(((((((.((....))(((.(((....))).)))..................)))))))..))..))..........)).------.....------. ( -31.30) >DroGri_CAF1 108491 103 + 1 GGGCGUGCAGCUGCAACAACAGGAGCUGCAGCAACAGCGCGGCGGCCUGCCGGUGCCCCCACAAGAUAUCUCUAGUAGUUCAGGUCCGCAAACAUGAAAU-----------GA------A ((((.((.((((((.....((((.(((((.((....)))))))..))))...(((....)))............)))))))).)))).............-----------..------. ( -34.00) >DroWil_CAF1 127607 116 + 1 GGGCGUCCAAUUGCAACAACAGGAGCUGCAGCAGCAACGUGGCGGUCUACCAGUGCCCACACAAGAUAUAUCUAGUAGCUCAGGUCCGCCAU---UU-ACCAGCAUUUAAUUGCAAUCAA .((((.((..(((......)))(((((((.........(((..(((........)))..))).(((....))).))))))).))..))))..---..-....(((......)))...... ( -29.80) >DroMoj_CAF1 127707 103 + 1 GGGCGUGCAGCUGCAACAACAGGAGCUGCAGCAGCAGCGCGGCGGCCUGCCGGUGCCCACACAAGAUAUCUCUAGUAGUUCAGGUCCGCAAAAAAUUAAC-----------AA------A ((((.((.((((((.......((.(((((....)))))(((.(((....))).))))).....(((....))).)))))))).)))).............-----------..------. ( -36.60) >DroAna_CAF1 88851 98 + 1 GGGCGUUCAGCUGCAACAACAGGAGCUGCAACAGCAACGCGGAGGGCUGCCGGUGCCCACCCAAGAUAUAUCUAGUAGCUCAGGUCCGACAA---GUCGCC------------------- .(((((((((((.(.......).))))).)))......((((((((((((.(((....)))..(((....))).)))))))...)))).)..---...)))------------------- ( -31.00) >consensus GGGCGUCCAACUGCAACAACAGGAGCUGCAGCAGCAGCGCGGCGGCCUGCCGGUGCCCACACAAGAUAUAUCUAGUAGCUCAGGUCCGCAAA___GUAACC__________AA______A ((((......(((......)))(((((((.........(((.(((....))).))).......(((....))).)))))))..))))................................. (-25.20 = -24.95 + -0.25)

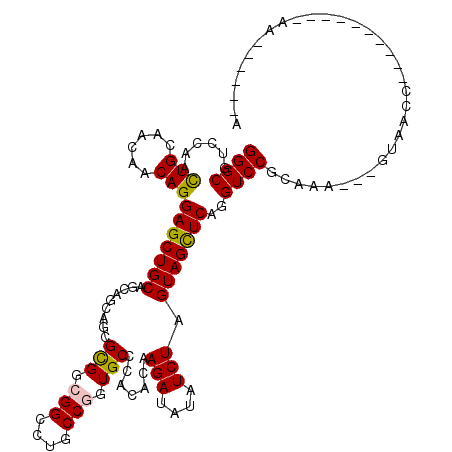
| Location | 10,974,047 – 10,974,152 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.15 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -25.01 |
| Energy contribution | -24.93 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10974047 105 - 20766785 ------GCAACU------UGGCGAC---UUGUCGGACCUGAGCUACUAGAUAUAUCUUGGCUGGGCACCGGCAGUCCGCCGCGCUGCUGCUGCAGCUCCUGUUGCUGCAGUUGUACGCCC ------......------.((((((---(.(((((..((.(((((..(((....)))))))).))..)))))))).))))(((.(((.((((((((.......)))))))).)))))).. ( -47.90) >DroVir_CAF1 111960 108 - 1 U------UUUUU------UGGUUUUUUUUUUGCGGACCUGAGCUACUAGAGAUAUCUUGUGUGGGCACCGGCAGACCGCCGCGCUGCUGCUGCAGCUCCUGUUGCUGCAAUUGGACGCCC .------.....------.(((.(((...((((((....(((((((.(((....))).))....(((.((((((.(......))))))).))))))))......))))))..))).))). ( -31.60) >DroGri_CAF1 108491 103 - 1 U------UC-----------AUUUCAUGUUUGCGGACCUGAACUACUAGAGAUAUCUUGUGGGGGCACCGGCAGGCCGCCGCGCUGUUGCUGCAGCUCCUGUUGUUGCAGCUGCACGCCC .------..-----------...........((((.((((..((((.(((....))).))))((...))..)))))))).(((.(((.((((((((.......)))))))).)))))).. ( -40.10) >DroWil_CAF1 127607 116 - 1 UUGAUUGCAAUUAAAUGCUGGU-AA---AUGGCGGACCUGAGCUACUAGAUAUAUCUUGUGUGGGCACUGGUAGACCGCCACGUUGCUGCUGCAGCUCCUGUUGUUGCAAUUGGACGCCC (..((((((((.....((.((.-..---.(((((((((.(.(((((.(((....)))...)).))).).)))...)))))).(((((....))))).)).)).))))))))..)...... ( -37.00) >DroMoj_CAF1 127707 103 - 1 U------UU-----------GUUAAUUUUUUGCGGACCUGAACUACUAGAGAUAUCUUGUGUGGGCACCGGCAGGCCGCCGCGCUGCUGCUGCAGCUCCUGUUGUUGCAGCUGCACGCCC .------..-----------...........((((.((((..((((.(((....)))...)))).......)))))))).(((.(((.((((((((.......)))))))).)))))).. ( -40.30) >DroAna_CAF1 88851 98 - 1 -------------------GGCGAC---UUGUCGGACCUGAGCUACUAGAUAUAUCUUGGGUGGGCACCGGCAGCCCUCCGCGUUGCUGUUGCAGCUCCUGUUGUUGCAGCUGAACGCCC -------------------((((..---..(.((((((..((.((.......)).))..)))((((.......)))).))))...(((((.(((((....))))).)))))....)))). ( -37.10) >consensus U______UA__________GGUUAC___UUGGCGGACCUGAGCUACUAGAGAUAUCUUGUGUGGGCACCGGCAGACCGCCGCGCUGCUGCUGCAGCUCCUGUUGUUGCAGCUGCACGCCC ...........................................(((.(((....))).))).((((..((((.....))))....(((((.(((((....))))).))))).....)))) (-25.01 = -24.93 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:10 2006