
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,961,145 – 10,961,323 |
| Length | 178 |
| Max. P | 0.796244 |

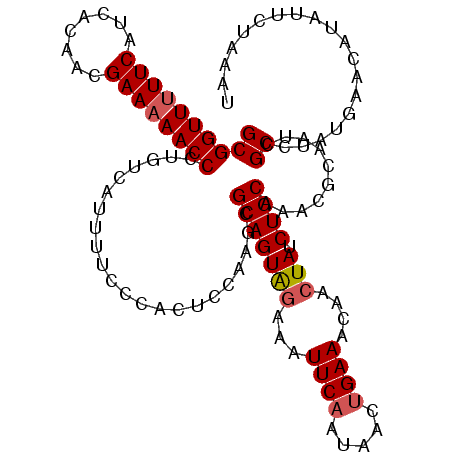
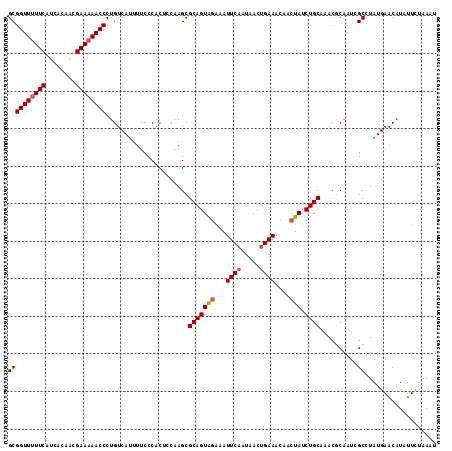
| Location | 10,961,145 – 10,961,259 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 92.37 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -16.52 |
| Energy contribution | -16.96 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
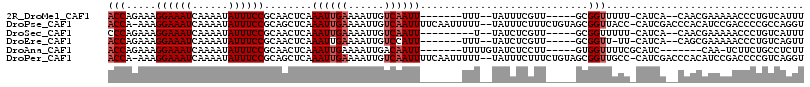
>2R_DroMel_CAF1 10961145 114 - 20766785 GCGGUUUUUCAUCACAACGAAAAACCCUGUCAUUUUCCCACUCCAAGCGCAGUAGAAAUUCAAUAACUGAAACCACUAUCUGCAAACGCAAUCGCCUAUGAACCUAUUCUAAAU ((((((((((........))))))))......................(((((((...((((.....))))....))).))))....))......................... ( -19.30) >DroSec_CAF1 73500 114 - 1 GCGGUUUUUCAUCACAACGAAAAACCCUGUCAUUUUCCCACUCCAAGCGCAGUAGGAAUUCAAUAACUGAAACAACUAUCUGCAAACGCAAUGGCCUAUGAACAUAUUCUAAAU ..((((((((........)))))))).((((((....(((........(((((((...((((.....))))....))).))))........)))...))).))).......... ( -21.19) >DroSim_CAF1 75777 114 - 1 GCGGUUUUUCAUCACAACGAAAAACCCUGUCAUUUUCCCACUCCAAGCGCAGUAGGAAUUCAAUAACUGAAACCACUAUCUGCAAACGCAAUCGCCUAUGAACAUAUUCUAAAC ((((((((((........))))))))......................((((((((..((((.....))))..).))).))))....))......................... ( -20.40) >DroEre_CAF1 75810 113 - 1 GCGGUU-UUCAUCACAGCGAAAAACCCUGUCAGUUUCCCACACCAAGCGCAGUGGAAAUUCAAUAACUGAAACGACUAUCUGCUAACACAAUCGCCUAUGAACAUGUUUUAAAU (((...-(((((....((((............((((........))))(((((((...((((.....))))....))).))))........))))..)))))..)))....... ( -15.80) >DroYak_CAF1 76925 114 - 1 GCGGUUUUUCAUCACAACGAAAAACCCUGUCAUAUUCCCACACCGAGCGCAGUAGAAAUUCCAUAACUGAAACAAAUAUCUGCCAACACAAUCGCCUAUGAACAUAUUUUAAAU ((((((((((........))))))))....................(.((((((....(((.......))).....)).))))).........))................... ( -13.80) >consensus GCGGUUUUUCAUCACAACGAAAAACCCUGUCAUUUUCCCACUCCAAGCGCAGUAGAAAUUCAAUAACUGAAACAACUAUCUGCAAACGCAAUCGCCUAUGAACAUAUUCUAAAU ((((((((((........))))))))......................(((((((...((((.....))))....))).))))..........))................... (-16.52 = -16.96 + 0.44)

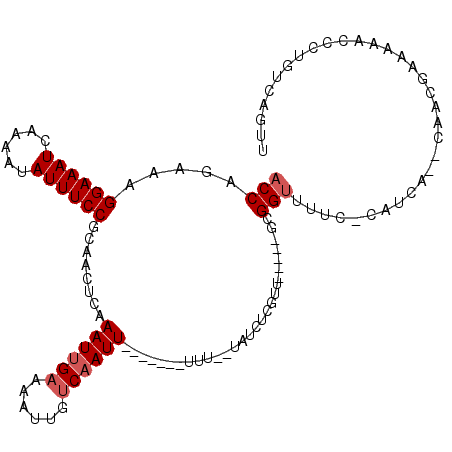
| Location | 10,961,224 – 10,961,323 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -7.85 |
| Energy contribution | -8.35 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10961224 99 - 20766785 ACCAGAAAGGAAAUCAAAAUAUUUCCGCAACUCAAAUUGAAAAUUGUCAAUU-------UUU--UAUUUCGUU-----GCGGUUUUU-CAUCA--CAACGAAAAACCCUGUCAUUU ..(((...((((((......))))))(((((..((((.((((((.....)))-------)))--.)))).)))-----))(((((((-(....--....)))))))))))...... ( -24.00) >DroPse_CAF1 68427 112 - 1 ACCA-AAAGGAAAUCAAAAUAUUUCCGCAGCUCAAAUUGAAAAUUGUCAAUUUUCAAUUUUU--UAUUUCUUUCUGUAGCGGUUACC-CAUCGACCCACAUCCGACCCCGCCAGGU (((.-...((((((......))))))((((...((((.((((((((........))))))))--.))))....)))).((((.....-..(((.........)))..))))..))) ( -20.40) >DroSec_CAF1 73579 97 - 1 CCCAGAAAGGAAAUCAAAAUAUUUCCGCAACUCAAAUUGAAAAUUGUCAAUU---------U--UAUCUCGUU-----GCGGUUUUU-CAUCA--CAACGAAAAACCCUGUCAUUU ..(((...((((((......))))))(((((..(((((((......))))))---------)--......)))-----))(((((((-(....--....)))))))))))...... ( -23.90) >DroEre_CAF1 75889 98 - 1 ACCAGAAAGGAAAUCAAAAUAUUUCCGCAACUCAAAUUGAAAAUUGUCCAUU-------UUU--UAUCUCGUU-----GCGGUU-UU-CAUCA--CAGCGAAAAACCCUGUCAGUU ....((.(((..............(((((((......((((((........)-------)))--))....)))-----))))((-((-(....--....)))))..))).)).... ( -18.30) >DroAna_CAF1 73858 96 - 1 ACCAGAAAGGAAAUCAAAAUAUUUCCGCAACUCAAAUUGAAAAUUGACAAUU-------UUUUGUAUCUCCUU-----GUGGUUUUCGCAUC-------CAA-UCUUCUGCCUCUU ..(((((.((((((......))))))((.((..((((((........)))))-------)...))....((..-----..)).....))...-------...-..)))))...... ( -14.10) >DroPer_CAF1 69093 112 - 1 ACCA-AAAGGAAAUCAAAAUAUUUCCGCAGCUCAAAUUGAAAAUUGUCAAUUUUCAAUUUUU--UAUUUCUUUCUGUAGCGGUUGCC-CAUCGACCCACAUCCGACCCCGUCAGGU (((.-...((((((......))))))((((...((((.((((((((........))))))))--.))))....)))).(.(((((..-...))))))......(((...))).))) ( -21.00) >consensus ACCAGAAAGGAAAUCAAAAUAUUUCCGCAACUCAAAUUGAAAAUUGUCAAUU_______UUU__UAUCUCGUU_____GCGGUUUUC_CAUCA__CAACGAAAAACCCUGUCAGUU (((.....((((((......))))))........((((((......))))))............................)))................................. ( -7.85 = -8.35 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:04 2006