| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,958,492 – 10,958,585 |
| Length | 93 |
| Max. P | 0.874553 |

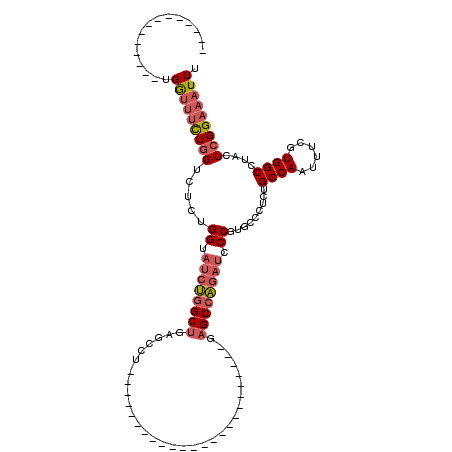
| Location | 10,958,492 – 10,958,585 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.09 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -15.34 |
| Energy contribution | -15.84 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10958492 93 + 20766785 AAAUUUCCGAGUAGGCCACGAAAUUGGCAGGGGGCACGGGAUCUGGCUCCGGCUC------------AGGCUGAGGCUCAGCCAGAUACCAGAGACCAGAAACCA-------------- ...(..((......((((......)))).))..)...((..((((((((.((.((------------.(((((.....))))).))..)).))).)))))..)).-------------- ( -35.70) >DroPse_CAF1 66654 71 + 1 AAAUUUCCGAGUAGGCCACGAAAUUGGCAAGGCACUC------CCGCUCC-----------------------AGGUCCUGCCGCUU-----CGACUAGUCUUCC-------------- ........((.(((((((......))))..((((...------((.....-----------------------.))...))))....-----...))).))....-------------- ( -18.30) >DroSec_CAF1 70887 81 + 1 AAAUUUCCGAGUAGGCCACGAAAUUGGCAGAGGACACGGGAUCUGGCUC------------------------AGGCUCAGCCAGAUACCAGAGACCAAAAACCA-------------- .....(((......((((......))))...)))...((..(((((.((------------------------.(((...))).))..)))))..))........-------------- ( -23.10) >DroSim_CAF1 73180 81 + 1 AAAUUUCCGAGUAGGCCACGAAAUUGGCAGAGGGCACGGGAUCUGGCUC------------------------AGGCUCAGCCAGAUACCAGAGACCAAAAACCA-------------- ...((((((.((..((((......)))).....)).))((((((((((.------------------------......)))))))).)).))))..........-------------- ( -22.80) >DroEre_CAF1 73210 105 + 1 AAAUUUCCGAGUAGGCCACGAAAUUGGCAGAGGGCACGGGAUCUGGCUCAGGCUCCGGCUCCGGCUCCGGCUGAGGCUCAGCCAGAUACCAGUAACCAAAAACCA-------------- ..............((((......))))...((..((.((((((((((..(((..((((..((....))))))..))).)))))))).)).))..))........-------------- ( -37.80) >DroYak_CAF1 74161 107 + 1 AAAUUUCCGAGUAGGCCACGAAAUUGGCAGAGGCCACGGGAUCUGGCUCACGCUC------------AGGCUGAGGCUCAGCCAGAUACCAGAUACCAGAAACCAGCAACCAAAAAACC ...((((.(.(((((((.(..........).))))..((.((((((((....(((------------.....)))....)))))))).))...)))).))))................. ( -28.70) >consensus AAAUUUCCGAGUAGGCCACGAAAUUGGCAGAGGGCACGGGAUCUGGCUC________________________AGGCUCAGCCAGAUACCAGAGACCAAAAACCA______________ ..............((((......))))...((.....(((((((((.................................))))))).)).....))...................... (-15.34 = -15.84 + 0.50)

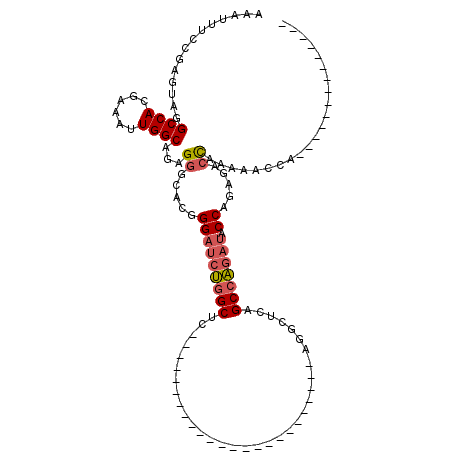
| Location | 10,958,492 – 10,958,585 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.09 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -17.55 |
| Energy contribution | -18.46 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10958492 93 - 20766785 --------------UGGUUUCUGGUCUCUGGUAUCUGGCUGAGCCUCAGCCU------------GAGCCGGAGCCAGAUCCCGUGCCCCCUGCCAAUUUCGUGGCCUACUCGGAAAUUU --------------.((..(((((.(((((((.((.(((((.....))))).------------))))))))))))))..))...((....((((......))))......))...... ( -39.50) >DroPse_CAF1 66654 71 - 1 --------------GGAAGACUAGUCG-----AAGCGGCAGGACCU-----------------------GGAGCGG------GAGUGCCUUGCCAAUUUCGUGGCCUACUCGGAAAUUU --------------....((.(((.((-----(((.((((((.(((-----------------------(...)))------)....)).))))..)))))....))).))........ ( -15.90) >DroSec_CAF1 70887 81 - 1 --------------UGGUUUUUGGUCUCUGGUAUCUGGCUGAGCCU------------------------GAGCCAGAUCCCGUGUCCUCUGCCAAUUUCGUGGCCUACUCGGAAAUUU --------------((((....((.(.(.((.((((((((......------------------------.)))))))))).).).))...)))).(((((.(.....).))))).... ( -27.10) >DroSim_CAF1 73180 81 - 1 --------------UGGUUUUUGGUCUCUGGUAUCUGGCUGAGCCU------------------------GAGCCAGAUCCCGUGCCCUCUGCCAAUUUCGUGGCCUACUCGGAAAUUU --------------((((....(((...(((.((((((((......------------------------.)))))))).))).)))....)))).(((((.(.....).))))).... ( -26.30) >DroEre_CAF1 73210 105 - 1 --------------UGGUUUUUGGUUACUGGUAUCUGGCUGAGCCUCAGCCGGAGCCGGAGCCGGAGCCUGAGCCAGAUCCCGUGCCCUCUGCCAAUUUCGUGGCCUACUCGGAAAUUU --------------((((....((.(((.((.((((((((.((.(((..(((....))).....))).)).)))))))))).))).))...)))).(((((.(.....).))))).... ( -41.50) >DroYak_CAF1 74161 107 - 1 GGUUUUUUGGUUGCUGGUUUCUGGUAUCUGGUAUCUGGCUGAGCCUCAGCCU------------GAGCGUGAGCCAGAUCCCGUGGCCUCUGCCAAUUUCGUGGCCUACUCGGAAAUUU (((.....(((..(.((..((((((.((.....((.(((((.....))))).------------))....))))))))..)))..)))...)))..(((((.(.....).))))).... ( -39.70) >consensus ______________UGGUUUCUGGUCUCUGGUAUCUGGCUGAGCCU________________________GAGCCAGAUCCCGUGCCCUCUGCCAAUUUCGUGGCCUACUCGGAAAUUU ...............(((((((((.....((.((((((((...............................)))))))).)).........((((......))))....))))))))). (-17.55 = -18.46 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:01 2006