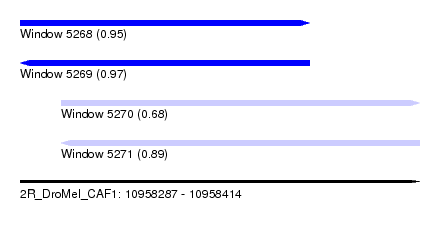
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,958,287 – 10,958,414 |
| Length | 127 |
| Max. P | 0.966641 |

| Location | 10,958,287 – 10,958,379 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -15.06 |
| Consensus MFE | -10.80 |
| Energy contribution | -11.49 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10958287 92 + 20766785 ---UUUUGGU----------CUC---------GCUUUUUUUUUAUCAAAAGCAAUUUUGCAC-UUUCUUUGGCUUGGCCUUUUAAUAAAUUCCACCAAGCUUUGCAUUUUCAGCA ---..((((.----------...---------((((((........)))))).....((((.-.......(((((((.................))))))).))))...)))).. ( -14.53) >DroPse_CAF1 66485 82 + 1 ---UUUCGUU-------------------------U----UUUAUCAAAAGCAAUUUUGCAA-UUUCUUUUGCUUGGCCUUUUAAUAAAUUCCGCCAAGCUUUGCAUUUUCAGCA ---....(((-------------------------(----((....)))))).....(((((-........(((((((...............))))))).)))))......... ( -15.56) >DroWil_CAF1 108740 112 + 1 GAAUUUUUUCAAUUUGCUUCUUCAUUUUACUUUUUUU---UUUAUCAAAAGCAAUUUUGCAAUUUUCUUUGGCUUGGCCUUUUAAUAAAUUCCGGCGAGCUUUGCAUUUUCAGCA (((..........((((((..................---........))))))...(((((........((((((.((..............)))))))))))))..))).... ( -13.91) >DroYak_CAF1 73943 95 + 1 ---UUUCGUUC----UCGUUCUC---------GCUUU---UUUAUCAAAAGCAAUUUUGCAC-UUUCUUUGGCUUGGCCUUUUAAUAAAUUCCACCAAGCUUUGCAUUUUCAGCA ---........----..(((...---------(((((---(.....)))))).....((((.-.......(((((((.................))))))).)))).....))). ( -14.83) >DroAna_CAF1 71199 104 + 1 ---UUUUGCCA----GCGUUUUCCUCUUCUUUUUUUU---UUUAUCAAAAGCAAUUUUGCAC-UUUCUUUGGCUUGGCCUUUUAAUAAAUUCCGCCAAGCUUUGCAUUUUCAGCA ---........----((....................---.....(((((....)))))...-.......((((((((...............))))))))...........)). ( -15.96) >DroPer_CAF1 67157 83 + 1 ---UUUCGUU-------------------------UU---UUUAUCAAAAGCAAUUUUGCAA-UUUCUUUUGCUUGGCCUUUUAAUAAAUUCCGCCAAGCUUUGCAUUUUCAGCA ---....(((-------------------------((---(.....)))))).....(((((-........(((((((...............))))))).)))))......... ( -15.56) >consensus ___UUUCGUU___________UC___________UUU___UUUAUCAAAAGCAAUUUUGCAA_UUUCUUUGGCUUGGCCUUUUAAUAAAUUCCGCCAAGCUUUGCAUUUUCAGCA ..................................................((.....((((.........((((((((...............)))))))).))))......)). (-10.80 = -11.49 + 0.70)

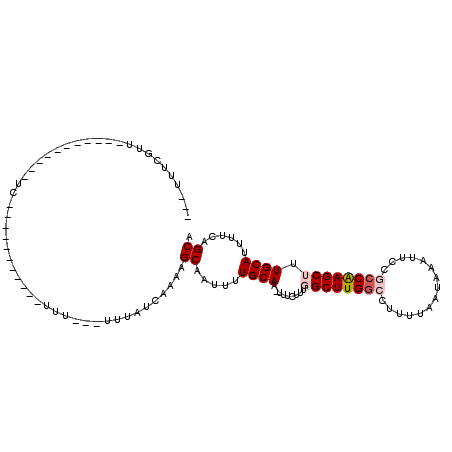
| Location | 10,958,287 – 10,958,379 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -17.43 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.11 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10958287 92 - 20766785 UGCUGAAAAUGCAAAGCUUGGUGGAAUUUAUUAAAAGGCCAAGCCAAAGAAA-GUGCAAAAUUGCUUUUGAUAAAAAAAAAGC---------GAG----------ACCAAAA--- .........((((...((((((((..(((....)))..))..))).)))...-.))))...((((((((........))))))---------)).----------.......--- ( -18.60) >DroPse_CAF1 66485 82 - 1 UGCUGAAAAUGCAAAGCUUGGCGGAAUUUAUUAAAAGGCCAAGCAAAAGAAA-UUGCAAAAUUGCUUUUGAUAAA----A-------------------------AACGAAA--- (((.......)))..(((((((...............)))))))(((((.((-((....)))).)))))......----.-------------------------.......--- ( -16.56) >DroWil_CAF1 108740 112 - 1 UGCUGAAAAUGCAAAGCUCGCCGGAAUUUAUUAAAAGGCCAAGCCAAAGAAAAUUGCAAAAUUGCUUUUGAUAAA---AAAAAAAGUAAAAUGAAGAAGCAAAUUGAAAAAAUUC ((((.....(((((.(((.(((..............)))..))).........)))))...((((((((......---...))))))))........)))).............. ( -16.04) >DroYak_CAF1 73943 95 - 1 UGCUGAAAAUGCAAAGCUUGGUGGAAUUUAUUAAAAGGCCAAGCCAAAGAAA-GUGCAAAAUUGCUUUUGAUAAA---AAAGC---------GAGAACGA----GAACGAAA--- .........((((...((((((((..(((....)))..))..))).)))...-.))))...((((((((.....)---)))))---------))......----........--- ( -18.60) >DroAna_CAF1 71199 104 - 1 UGCUGAAAAUGCAAAGCUUGGCGGAAUUUAUUAAAAGGCCAAGCCAAAGAAA-GUGCAAAAUUGCUUUUGAUAAA---AAAAAAAAGAAGAGGAAAACGC----UGGCAAAA--- ((((...........(((((((...............))))))).......(-(((........(((((......---.......))))).......)))----)))))...--- ( -18.24) >DroPer_CAF1 67157 83 - 1 UGCUGAAAAUGCAAAGCUUGGCGGAAUUUAUUAAAAGGCCAAGCAAAAGAAA-UUGCAAAAUUGCUUUUGAUAAA---AA-------------------------AACGAAA--- (((.......)))..(((((((...............)))))))(((((.((-((....)))).)))))......---..-------------------------.......--- ( -16.56) >consensus UGCUGAAAAUGCAAAGCUUGGCGGAAUUUAUUAAAAGGCCAAGCCAAAGAAA_GUGCAAAAUUGCUUUUGAUAAA___AAA___________GA___________AACAAAA___ (((.......)))..(((((((...............)))))))...........(((....))).................................................. (-12.00 = -12.11 + 0.11)
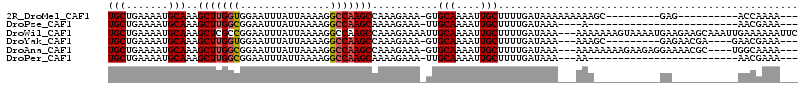
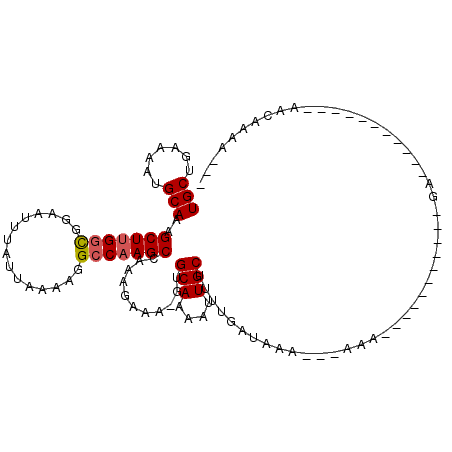

| Location | 10,958,300 – 10,958,414 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -18.96 |
| Consensus MFE | -11.50 |
| Energy contribution | -11.86 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.680693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10958300 114 + 20766785 UUUUUUUUAUCAAAAGCAAUUUUGCAC-UUUCUUUGGCUUGGCCUUUUAAUAAAUUCCACCAAGCUUUGCAUUUUCAGCAGUUUUCC--GGGC--AGCAUUUCUCCGCC-CGGCUAUUCU ..........(((((....)))))...-.......(((((((.................)))))))((((.......))))....((--((((--...........)))-)))....... ( -23.13) >DroPse_CAF1 66492 92 + 1 U----UUUAUCAAAAGCAAUUUUGCAA-UUUCUUUUGCUUGGCCUUUUAAUAAAUUCCGCCAAGCUUUGCAUUUUCAGCAGUUUUCG--CUGU--G---GUUC----------------G .----...((((..........(((((-........(((((((...............))))))).)))))....((((.......)--))))--)---))..----------------. ( -17.86) >DroWil_CAF1 108775 93 + 1 UU---UUUAUCAAAAGCAAUUUUGCAAUUUUCUUUGGCUUGGCCUUUUAAUAAAUUCCGGCGAGCUUUGCAUUUUCAGCAGUUUCCG--UU-U--G---CUUC----------------U ..---........((((((....((..........((((((.((..............)))))))).(((.......))))).....--.)-)--)---))).----------------. ( -14.74) >DroYak_CAF1 73962 112 + 1 UU---UUUAUCAAAAGCAAUUUUGCAC-UUUCUUUGGCUUGGCCUUUUAAUAAAUUCCACCAAGCUUUGCAUUUUCAGCAGUUUUCC--GGGC--AGCAUUUCUCCGCCCCGGUUAUUCU ..---.....(((((....)))))...-.......(((((((.................)))))))((((.......))))....((--(((.--.((........)))))))....... ( -21.53) >DroAna_CAF1 71227 102 + 1 UU---UUUAUCAAAAGCAAUUUUGCAC-UUUCUUUGGCUUGGCCUUUUAAUAAAUUCCGCCAAGCUUUGCAUUUUCAGCAGUUUUUCCUCGGCAAAACAUUUCUCC-------------- ..---..............((((((..-.......((((((((...............))))))))((((.......))))..........)))))).........-------------- ( -18.66) >DroPer_CAF1 67164 93 + 1 UU---UUUAUCAAAAGCAAUUUUGCAA-UUUCUUUUGCUUGGCCUUUUAAUAAAUUCCGCCAAGCUUUGCAUUUUCAGCAGUUUUCG--CUGU--G---GUUC----------------G ..---...((((..........(((((-........(((((((...............))))))).)))))....((((.......)--))))--)---))..----------------. ( -17.86) >consensus UU___UUUAUCAAAAGCAAUUUUGCAA_UUUCUUUGGCUUGGCCUUUUAAUAAAUUCCGCCAAGCUUUGCAUUUUCAGCAGUUUUCC__CGGC__A___UUUC________________U ...............(((....)))...........(((((((...............))))))).((((.......))))....................................... (-11.50 = -11.86 + 0.36)
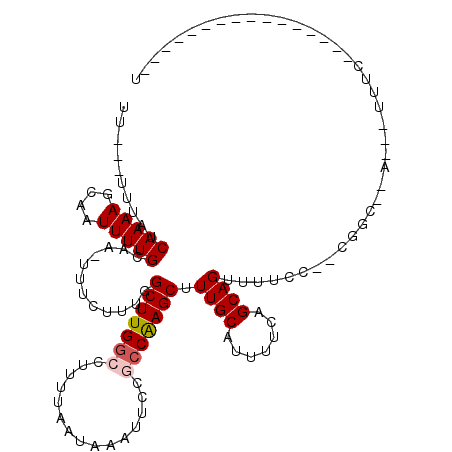
| Location | 10,958,300 – 10,958,414 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -12.10 |
| Energy contribution | -12.21 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10958300 114 - 20766785 AGAAUAGCCG-GGCGGAGAAAUGCU--GCCC--GGAAAACUGCUGAAAAUGCAAAGCUUGGUGGAAUUUAUUAAAAGGCCAAGCCAAAGAAA-GUGCAAAAUUGCUUUUGAUAAAAAAAA .......(((-((((((....).))--))))--)).....(((.......)))..(((((((...............)))))))....((((-((........))))))........... ( -30.56) >DroPse_CAF1 66492 92 - 1 C----------------GAAC---C--ACAG--CGAAAACUGCUGAAAAUGCAAAGCUUGGCGGAAUUUAUUAAAAGGCCAAGCAAAAGAAA-UUGCAAAAUUGCUUUUGAUAAA----A .----------------....---.--.(((--((.....)))))..........(((((((...............)))))))(((((.((-((....)))).)))))......----. ( -20.16) >DroWil_CAF1 108775 93 - 1 A----------------GAAG---C--A-AA--CGGAAACUGCUGAAAAUGCAAAGCUCGCCGGAAUUUAUUAAAAGGCCAAGCCAAAGAAAAUUGCAAAAUUGCUUUUGAUAAA---AA (----------------((((---(--(-(.--(((.....(((..........)))...))).............(((...)))................))))))))......---.. ( -16.80) >DroYak_CAF1 73962 112 - 1 AGAAUAACCGGGGCGGAGAAAUGCU--GCCC--GGAAAACUGCUGAAAAUGCAAAGCUUGGUGGAAUUUAUUAAAAGGCCAAGCCAAAGAAA-GUGCAAAAUUGCUUUUGAUAAA---AA .......(((((.((((....).))--))))--)).....(((.......)))..(((((((...............)))))))....((((-((........))))))......---.. ( -27.06) >DroAna_CAF1 71227 102 - 1 --------------GGAGAAAUGUUUUGCCGAGGAAAAACUGCUGAAAAUGCAAAGCUUGGCGGAAUUUAUUAAAAGGCCAAGCCAAAGAAA-GUGCAAAAUUGCUUUUGAUAAA---AA --------------.........((((((((((.......(((.......)))...))))))))))(((((((((((((...))).......-..(((....)))))))))))))---.. ( -22.20) >DroPer_CAF1 67164 93 - 1 C----------------GAAC---C--ACAG--CGAAAACUGCUGAAAAUGCAAAGCUUGGCGGAAUUUAUUAAAAGGCCAAGCAAAAGAAA-UUGCAAAAUUGCUUUUGAUAAA---AA .----------------....---.--.(((--((.....)))))..........(((((((...............)))))))(((((.((-((....)))).)))))......---.. ( -20.16) >consensus A________________GAAA___C__ACAG__CGAAAACUGCUGAAAAUGCAAAGCUUGGCGGAAUUUAUUAAAAGGCCAAGCCAAAGAAA_GUGCAAAAUUGCUUUUGAUAAA___AA ........................................(((.......)))..(((((((...............)))))))...........(((....)))............... (-12.10 = -12.21 + 0.11)

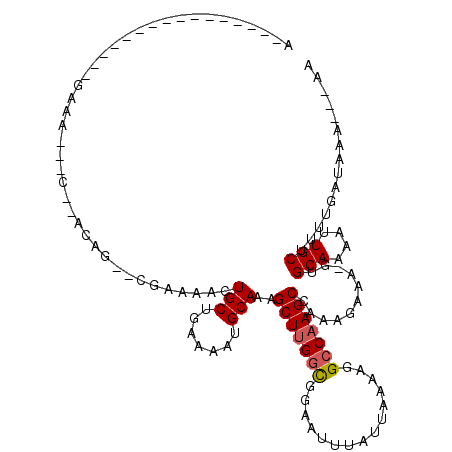
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:59 2006