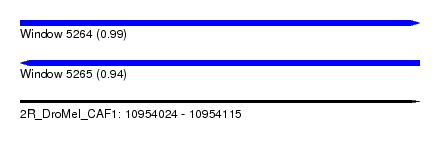
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,954,024 – 10,954,115 |
| Length | 91 |
| Max. P | 0.988823 |

| Location | 10,954,024 – 10,954,115 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -22.83 |
| Energy contribution | -22.95 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
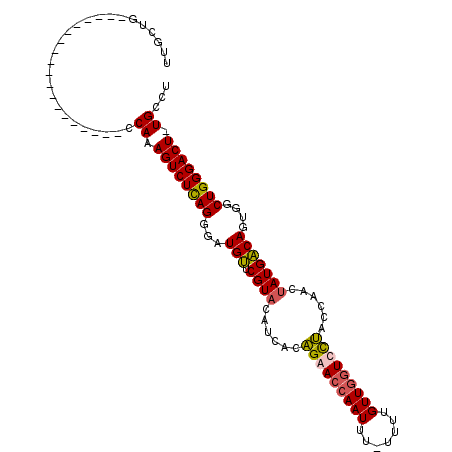
>2R_DroMel_CAF1 10954024 91 + 20766785 UUGCUG-------------------CCAAAGUCUCAGGGAUGUUCGUACAUCACAGAACCAAUUUUUUUUGUUGGUUCUACCAACUAUGACAGUGGCUGGGACUUUGCCU .....(-------------------.(((((((((((.(((((....)))))..(((((((((.......))))))))).(((.((.....)))))))))))))))).). ( -32.60) >DroSec_CAF1 66531 89 + 1 UUGCUG-------------------CCAAAGUCUUAGGGAUGUUCGUACAUCACAGAACCAAUUU-UUUUGUUGGUCUUACCAACUAUGACAGUGGCUGGGACU-UGCCU .....(-------------------(..(((((((((.(((((....)))))......(((....-....(((((.....)))))........)))))))))))-))).. ( -24.99) >DroSim_CAF1 68786 89 + 1 UUGCUG-------------------CCAAAGUCUCAGGGAUGUUCGUACAUCACAGAACCAAUUU-UUUUGUUGGUCCUACCAACUAUGACAGUGGCUGGGACU-UGCCU .....(-------------------(..(((((((((.(((((....)))))......(((....-....(((((.....)))))........)))))))))))-))).. ( -27.09) >DroEre_CAF1 68646 106 + 1 AGGG--UCUUGUUACAAAGGCAAAGCCAAAGUCUCAGGCAUGCUCGUACAUCAGGGAACCAAUUU-UUUCGUUAAUUCCACCAACGAUGGCAGUGACUGGGACU-UGCCU ..((--..(((((.....)))))..)).(((((((((.(((((.(((.......((((..(((..-....)))..)))).......)))))).)).))))))))-).... ( -29.54) >consensus UUGCUG___________________CCAAAGUCUCAGGGAUGUUCGUACAUCACAGAACCAAUUU_UUUUGUUGGUCCUACCAACUAUGACAGUGGCUGGGACU_UGCCU ..........................((.((((((((...(((.((((......(((((((((.......)))))))))......)))))))....)))))))).))... (-22.83 = -22.95 + 0.12)

| Location | 10,954,024 – 10,954,115 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -17.19 |
| Energy contribution | -17.00 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.937974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
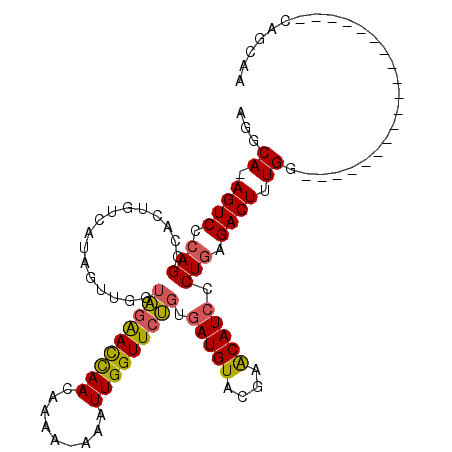
>2R_DroMel_CAF1 10954024 91 - 20766785 AGGCAAAGUCCCAGCCACUGUCAUAGUUGGUAGAACCAACAAAAAAAAUUGGUUCUGUGAUGUACGAACAUCCCUGAGACUUUGG-------------------CAGCAA .(.(((((((.(((((((((...))).)))(((((((((.........))))))))).(((((....))))).))).))))))).-------------------)..... ( -28.10) >DroSec_CAF1 66531 89 - 1 AGGCA-AGUCCCAGCCACUGUCAUAGUUGGUAAGACCAACAAAA-AAAUUGGUUCUGUGAUGUACGAACAUCCCUAAGACUUUGG-------------------CAGCAA .(((.-.......))).((((((.((((((((..(((((.....-...)))))..)).(((((....)))))))...)))).)))-------------------)))... ( -23.70) >DroSim_CAF1 68786 89 - 1 AGGCA-AGUCCCAGCCACUGUCAUAGUUGGUAGGACCAACAAAA-AAAUUGGUUCUGUGAUGUACGAACAUCCCUGAGACUUUGG-------------------CAGCAA .(((.-.......))).((((((.(((((((((((((((.....-...))))))))).(((((....)))))))...)))).)))-------------------)))... ( -28.40) >DroEre_CAF1 68646 106 - 1 AGGCA-AGUCCCAGUCACUGCCAUCGUUGGUGGAAUUAACGAAA-AAAUUGGUUCCCUGAUGUACGAGCAUGCCUGAGACUUUGGCUUUGCCUUUGUAACAAGA--CCCU (((((-((((.(((.((.((((..(((..(.((((((((.....-...)))))))))..)))...).))))).))).)))).......)))))...........--.... ( -28.61) >consensus AGGCA_AGUCCCAGCCACUGUCAUAGUUGGUAGAACCAACAAAA_AAAUUGGUUCUGUGAUGUACGAACAUCCCUGAGACUUUGG___________________CAGCAA ...((.((((.(((................(((((((((.........))))))))).(((((....))))).))).)))).)).......................... (-17.19 = -17.00 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:53 2006