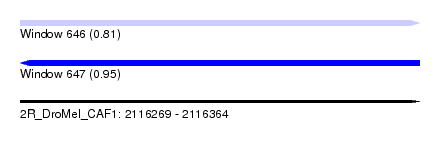
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,116,269 – 2,116,364 |
| Length | 95 |
| Max. P | 0.949960 |

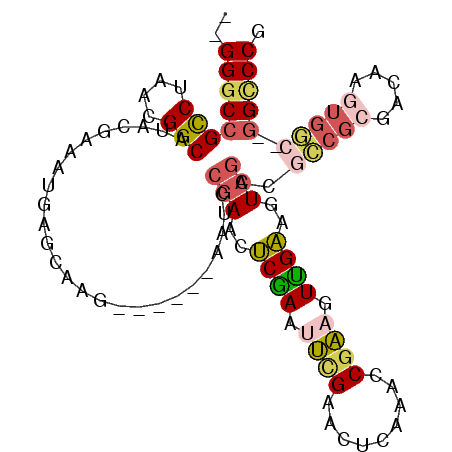
| Location | 2,116,269 – 2,116,364 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -17.40 |
| Energy contribution | -16.83 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
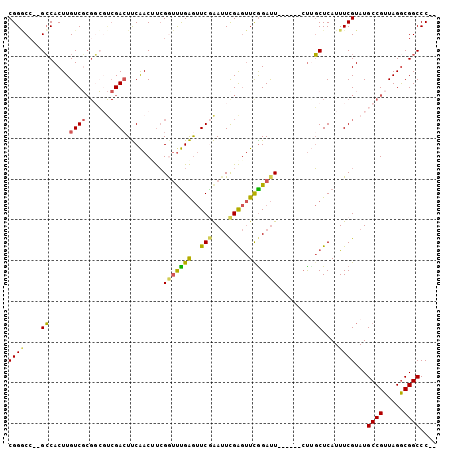
>2R_DroMel_CAF1 2116269 95 + 20766785 CGGGCC--GCCACUUGUCGCGGCGUCGACUUCAACUUCGGUUUGAGUUCGAAUUCGAGUUCGGAUU------CUUGCUCAUUUCGUAUGCCGUUAGGCGGCCC-- .(((((--(((.......((((((((((....(((....)))(((((..(((((((....))))))------)..)))))..))).)))))))..))))))))-- ( -47.00) >DroPse_CAF1 111025 105 + 1 CGGCCGGUGGCACUUGUCUCGCGGCCGACUCCGACUCGGACUCGGACUCGGACUCGGACUCGGACUACGAGUACGCCUCAUUUCGUAUGCCGUUAGGCGGCCCCG .((((.(.(((....((((((..(((((.(((((.((.((.......)).)).))))).)))).)..)))).))))).).........(((....)))))))... ( -43.20) >DroSec_CAF1 95009 95 + 1 CGGGCC--GCCACUUGUCGCGGCGUCGACUUCAACUUCGGUUUGAGUUCGAAUUCGAGUUUGGAUU------CUUGCUCAUUUCGUAUGCCGUUAGGCGGCCC-- .(((((--(((.......((((((((((....(((....)))(((((..(((((((....))))))------)..)))))..))).)))))))..))))))))-- ( -44.20) >DroSim_CAF1 98304 95 + 1 CGGGCC--GCCACUUGUCGCGGCGUCGACUUCAACUUCGGUUUGAGUUCGAAUUCGAGUUCGGAUU------CUUGCUCAUUUCGUAUGCCGUUAGGCGGCCC-- .(((((--(((.......((((((((((....(((....)))(((((..(((((((....))))))------)..)))))..))).)))))))..))))))))-- ( -47.00) >DroEre_CAF1 93329 87 + 1 CGGACC--GCCACUUGUC--------GACUUCAACAUCGGUCUGAGUUCAAAUUCGAGUUCAGAUU------CUUGCUCAUUUCGUAUGCCGUUAGGCGGCCC-- .((.((--(((((....(--------((......((..(((((((..((......))..)))))))------..))......)))......))..))))))).-- ( -23.80) >DroYak_CAF1 97053 95 + 1 CGGACC--GCCACUUGUCGCGGCCUCGAAUUCAACAUCGAUUUGAGUUCAAAUUUGAGUUCGGAUU------CUCGCUCAUUUCGUAUGCCGUUAGACGGCCC-- .((.((--((........)))))).............(((..(((((...((((((....))))))------...)))))..)))...((((.....))))..-- ( -27.30) >consensus CGGGCC__GCCACUUGUCGCGGCGUCGACUUCAACUUCGGUUUGAGUUCGAAUUCGAGUUCGGAUU______CUUGCUCAUUUCGUAUGCCGUUAGGCGGCCC__ ((((....((.....((((......)))).........(((((((..(((....)))..))))))).........))....))))...((((.....)))).... (-17.40 = -16.83 + -0.56)

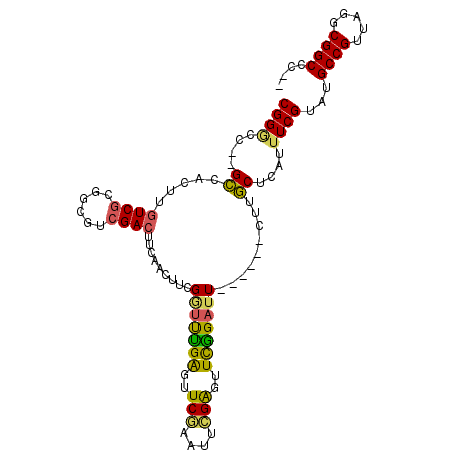
| Location | 2,116,269 – 2,116,364 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -24.59 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2116269 95 - 20766785 --GGGCCGCCUAACGGCAUACGAAAUGAGCAAG------AAUCCGAACUCGAAUUCGAACUCAAACCGAAGUUGAAGUCGACGCCGCGACAAGUGGC--GGCCCG --((((((((....)))...((...((((...(------(((.((....)).))))...))))...))..((((....))))(((((.....)))))--))))). ( -39.80) >DroPse_CAF1 111025 105 - 1 CGGGGCCGCCUAACGGCAUACGAAAUGAGGCGUACUCGUAGUCCGAGUCCGAGUCCGAGUCCGAGUCCGAGUCGGAGUCGGCCGCGAGACAAGUGCCACCGGCCG ...(((((((....)))...........(((((.((((..(.((((.(((((.((.((.......)).)).))))).)))))..))))))....)))...)))). ( -47.90) >DroSec_CAF1 95009 95 - 1 --GGGCCGCCUAACGGCAUACGAAAUGAGCAAG------AAUCCAAACUCGAAUUCGAACUCAAACCGAAGUUGAAGUCGACGCCGCGACAAGUGGC--GGCCCG --((((((((....)))...((...((((...(------(((.(......).))))...))))...))..((((....))))(((((.....)))))--))))). ( -37.00) >DroSim_CAF1 98304 95 - 1 --GGGCCGCCUAACGGCAUACGAAAUGAGCAAG------AAUCCGAACUCGAAUUCGAACUCAAACCGAAGUUGAAGUCGACGCCGCGACAAGUGGC--GGCCCG --((((((((....)))...((...((((...(------(((.((....)).))))...))))...))..((((....))))(((((.....)))))--))))). ( -39.80) >DroEre_CAF1 93329 87 - 1 --GGGCCGCCUAACGGCAUACGAAAUGAGCAAG------AAUCUGAACUCGAAUUUGAACUCAGACCGAUGUUGAAGUC--------GACAAGUGGC--GGUCCG --((((((((...((((((.....)))......------..(((((..(((....)))..)))))))).(((((....)--------))))...)))--))))). ( -30.40) >DroYak_CAF1 97053 95 - 1 --GGGCCGUCUAACGGCAUACGAAAUGAGCGAG------AAUCCGAACUCAAAUUUGAACUCAAAUCGAUGUUGAAUUCGAGGCCGCGACAAGUGGC--GGUCCG --((((((((....)))...((((.(.((((((------........)))..(((((....)))))....))).).))))..(((((.....)))))--))))). ( -31.60) >consensus __GGGCCGCCUAACGGCAUACGAAAUGAGCAAG______AAUCCGAACUCGAAUUCGAACUCAAACCGAAGUUGAAGUCGACGCCGCGACAAGUGGC__GGCCCG ..((((((((....)))..........................(((..((((.((((.........)))).))))..)))..(((((.....)))))..))))). (-24.59 = -25.40 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:21 2006