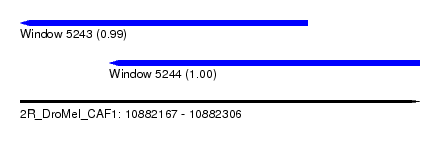
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,882,167 – 10,882,306 |
| Length | 139 |
| Max. P | 0.996718 |

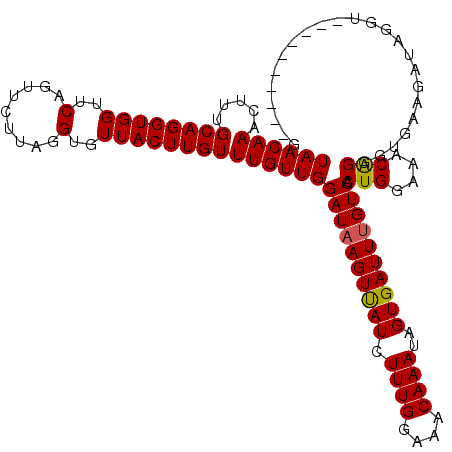
| Location | 10,882,167 – 10,882,267 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 86.32 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -21.29 |
| Energy contribution | -21.97 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10882167 100 - 20766785 UGUUUGUUGGAUUAGUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGUUAGGU----------AAAACUGGCAAAACUUUUAAAA-------ACUCAACGGA ..((((((((((.((((((.((((....))))..)))))).))).(((....)))(((...(((((..----------....)))))...))).......-------...))))))) ( -25.30) >DroSec_CAF1 13794 100 - 1 UGUUUGUUGGAUAAGUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGGUAGGU----------GAAACUGGCAAAACUUUUAAAA-------ACUCAACGGA ..(((((((((((((((((.((((....))))..)))))))))).(((....)))(((...(.(((..----------....))).)...))).......-------...))))))) ( -26.90) >DroSim_CAF1 10410 100 - 1 UGUUUGUUGGAUAAGUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGGUAGGU----------GAAACUGGCAAAACUUUUAAAA-------ACUCAACGGA ..(((((((((((((((((.((((....))))..)))))))))).(((....)))(((...(.(((..----------....))).)...))).......-------...))))))) ( -26.90) >DroEre_CAF1 10514 107 - 1 UGUUUGUUGGAUAAGUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGAUAGGU----------GAAACUGGGAAAACUUACAAAACGAAUACGUUCAACGGA ((((((((...((((((.((((((....))).((((.(((((((.(((....)))......)))))))----------...))))))).))))))...))))))))((.....)).. ( -26.80) >DroYak_CAF1 14485 115 - 1 UGUUUGUUGGAUAAGUCAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACGGGGUGAAGAUAGGUGAAGUUGGGUGAAACUGGGAAAACUUUCAAAACGGAUACAUUAAACU-- ((((((((.((((((((((.((((....))))..)))))))))).(((....)))..(((((........((((......))))........))))).)))))))).........-- ( -28.69) >consensus UGUUUGUUGGAUAAGUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGAUAGGU__________GAAACUGGCAAAACUUUUAAAA_______ACUCAACGGA ..(((((((((((((((((.((((....))))..)))))))))).(((....)))(((.....(((................))).....))).................))))))) (-21.29 = -21.97 + 0.68)

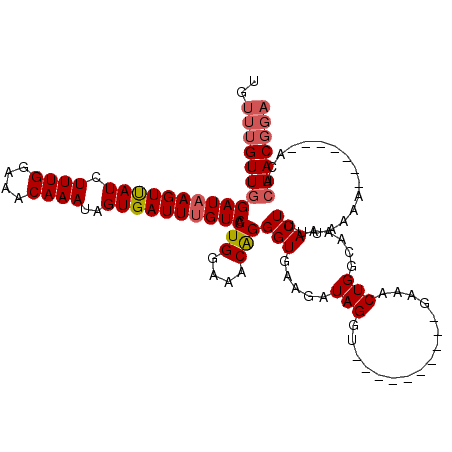
| Location | 10,882,198 – 10,882,306 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 94.29 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -26.54 |
| Energy contribution | -26.42 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10882198 108 - 20766785 UAACAAACUUUGCAGGUGGUUCAGUUCUUAGGUGUUACUUGUUUGUUGGAUUAGUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGUUAGGU----------A ...(.(((((..(..((..((((((...((((.....)))).......(((.((((((.((((....))))..)))))).))))))))).))...)..))))).)..----------. ( -24.90) >DroSec_CAF1 13825 108 - 1 UAACAAACUUUGCAGGUGGUUCAGUUCUUAGGUGUUACUUGUUUGUUGGAUAAGUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGGUAGGU----------G ..((..(((((((((((((..(.........)..))))))))......((((((((((.((((....))))..)))))))))).(((....)))....)))))..))----------. ( -27.00) >DroSim_CAF1 10441 108 - 1 UAACAAACUUUGCAGGUGGUUCAGUUCUUAGGUGUUACUUGUUUGUUGGAUAAGUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGGUAGGU----------G ..((..(((((((((((((..(.........)..))))))))......((((((((((.((((....))))..)))))))))).(((....)))....)))))..))----------. ( -27.00) >DroEre_CAF1 10552 108 - 1 UAACAAACUUUGCAGGUGGUUCAGUUCUUAGGUGUUACUUGUUUGUUGGAUAAGUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGAUAGGU----------G ((((((.....((((((((..(.........)..))))))))))))))((((((((((.((((....))))..)))))))))).(((....))).............----------. ( -26.50) >DroYak_CAF1 14521 118 - 1 UAACAAACUUUGCAGGUGGUUCAGUUCUUAGGUGUUACUUGUUUGUUGGAUAAGUCAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACGGGGUGAAGAUAGGUGAAGUUGGGUG ..((.(((((..(..((..((((.(((..(((.....)))((((.(..((((((((((.((((....))))..))))))))))...).))))))).)))).))..)..)))))..)). ( -31.50) >consensus UAACAAACUUUGCAGGUGGUUCAGUUCUUAGGUGUUACUUGUUUGUUGGAUAAGUUAUCUUUGGAAACAAAUAGUGAUUUGUCACUGGAAACAGGGUGAAGAUAGGU__________G ((((((.....((((((((..(.........)..))))))))))))))((((((((((.((((....))))..)))))))))).(((....)))........................ (-26.54 = -26.42 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:33 2006