
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
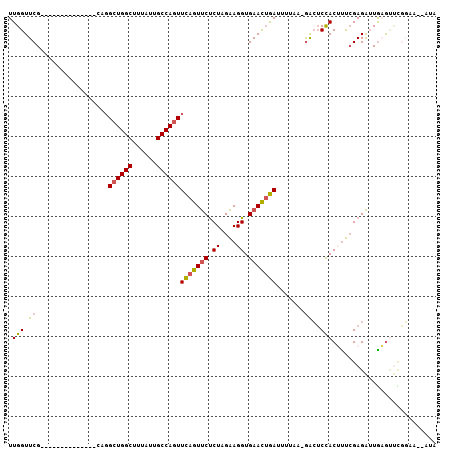
| Location | 10,879,044 – 10,879,134 |
| Length | 90 |
| Max. P | 0.986737 |

| Location | 10,879,044 – 10,879,134 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -18.33 |
| Energy contribution | -18.22 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10879044 90 + 20766785 UUGGUUCG--------------CAGGCUGGCUAUAUUGCCAGUUCAGUUCCCUAGAAGGUGAACUGAUUUUAA-GACUCCACUUUGGAGAUUGAAUUCGGAA--AUA ....((((--------------...((((((......))))))(((((((((.....)).))))))).(((((-..((((.....)))).)))))..)))).--... ( -29.10) >DroSec_CAF1 10702 90 + 1 UUGGUUCG--------------CAGGCUGGCUUUAUUGCCAGUUCAGUUCUCUAGAAGGUGAACUGAUUUUAA-GACUCCACUUUCGAGAUUGAGUUCGGAA--AUA .(((.((.--------------...((((((......))))))(((((((.((....)).)))))))......-))..))).(((((((......)))))))--... ( -27.30) >DroSim_CAF1 7370 80 + 1 UUGGUUCG--------------CAGGCUGGCUUUAUUGCCAGUUCAGUUCUCUAGAAGGUGAACUGAUUUUAA-GACUCCACUUUCGAGAUUGAG------------ .(((.((.--------------...((((((......))))))(((((((.((....)).)))))))......-))..)))..............------------ ( -22.90) >DroEre_CAF1 7429 92 + 1 UUGGUUCG--------------CAUGCUGGCUUUAUUGCCAGUUCAGUUCUCUAGAAGGUGAACUGAUUUGUAAGACUCUACGUUCGAGAUUACGUUUGGAG-UAUU .......(--------------((.((((((......))))))(((((((.((....)).)))))))..)))...(((((((((........))))..))))-)... ( -27.90) >DroYak_CAF1 11416 90 + 1 UUGGUUCG--------------CAGGCUGGCUUUAUUGCCAGUUCAGUUCUCUAGAAGGUGAACUGAUAUGUA-GACUCUACAUUCGAGAUUGCGUUUGGAA--AUA ((.(..((--------------(((((((((......))))))(((((((.((....)).))))))).(((((-(...))))))......)))))..).)).--... ( -28.70) >DroAna_CAF1 7205 105 + 1 UUGGUUCGGCAAGAAUUGCAAGCAGGCUGGCUUUAUUGCCA-UUUAGUCCUCUAGGAGGUGAAUGAAAUCCUA-AGGGCCACUCCCGAGGAUGAGCCCCCAAACAUA ..((((((((((.....((.((....)).))....))))).-....(((((((((((..(.....)..)))))-.(((.....)))))))))))))).......... ( -34.50) >consensus UUGGUUCG______________CAGGCUGGCUUUAUUGCCAGUUCAGUUCUCUAGAAGGUGAACUGAUUUUAA_GACUCCACUUUCGAGAUUGAGUUCGGAA__AUA .(((.((..................((((((......))))))(((((((.((....)).))))))).......))..))).......................... (-18.33 = -18.22 + -0.11)


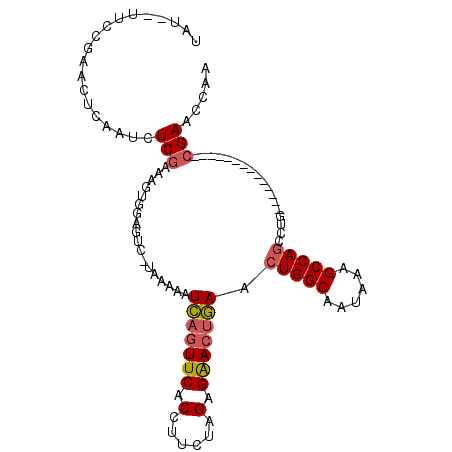
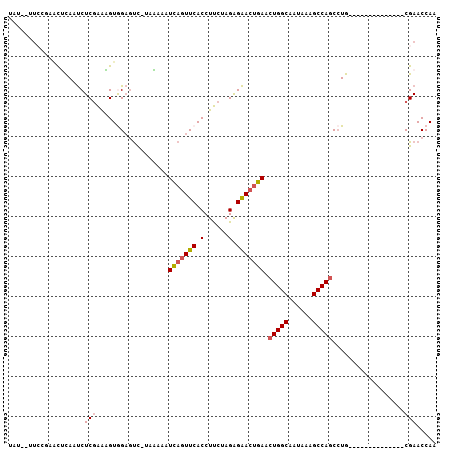
| Location | 10,879,044 – 10,879,134 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -14.66 |
| Energy contribution | -14.92 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10879044 90 - 20766785 UAU--UUCCGAAUUCAAUCUCCAAAGUGGAGUC-UUAAAAUCAGUUCACCUUCUAGGGAACUGAACUGGCAAUAUAGCCAGCCUG--------------CGAACCAA ...--.......(((...((((.....))))..-......(((((((.((.....))))))))).(((((......)))))....--------------.))).... ( -24.80) >DroSec_CAF1 10702 90 - 1 UAU--UUCCGAACUCAAUCUCGAAAGUGGAGUC-UUAAAAUCAGUUCACCUUCUAGAGAACUGAACUGGCAAUAAAGCCAGCCUG--------------CGAACCAA ...--....((.(((.....(....)..)))))-......(((((((.(......).))))))).(((((......)))))....--------------........ ( -20.30) >DroSim_CAF1 7370 80 - 1 ------------CUCAAUCUCGAAAGUGGAGUC-UUAAAAUCAGUUCACCUUCUAGAGAACUGAACUGGCAAUAAAGCCAGCCUG--------------CGAACCAA ------------.......(((.....((....-......(((((((.(......).))))))).(((((......)))))))..--------------)))..... ( -19.10) >DroEre_CAF1 7429 92 - 1 AAUA-CUCCAAACGUAAUCUCGAACGUAGAGUCUUACAAAUCAGUUCACCUUCUAGAGAACUGAACUGGCAAUAAAGCCAGCAUG--------------CGAACCAA ...(-(((...((((........)))).))))........(((((((.(......).))))))).(((((......)))))....--------------........ ( -20.60) >DroYak_CAF1 11416 90 - 1 UAU--UUCCAAACGCAAUCUCGAAUGUAGAGUC-UACAUAUCAGUUCACCUUCUAGAGAACUGAACUGGCAAUAAAGCCAGCCUG--------------CGAACCAA ...--.......((((.......((((((...)-))))).(((((((.(......).))))))).(((((......)))))..))--------------))...... ( -24.30) >DroAna_CAF1 7205 105 - 1 UAUGUUUGGGGGCUCAUCCUCGGGAGUGGCCCU-UAGGAUUUCAUUCACCUCCUAGAGGACUAAA-UGGCAAUAAAGCCAGCCUGCUUGCAAUUCUUGCCGAACCAA ..(((..(.((((...((((((((.....))).-(((((...........)))))))))).....-((((......)))))))).)..)))................ ( -31.90) >consensus UAU__UUCCGAACUCAAUCUCGAAAGUGGAGUC_UAAAAAUCAGUUCACCUUCUAGAGAACUGAACUGGCAAUAAAGCCAGCCUG______________CGAACCAA ...................(((..................(((((((.(......).))))))).(((((......)))))..................)))..... (-14.66 = -14.92 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:30 2006