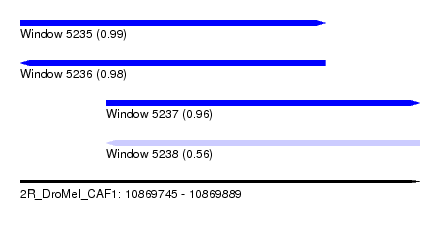
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,869,745 – 10,869,889 |
| Length | 144 |
| Max. P | 0.989810 |

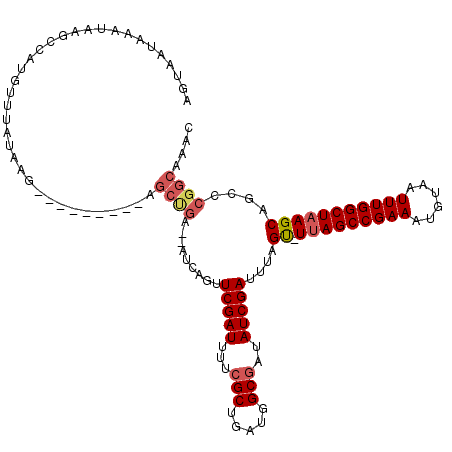
| Location | 10,869,745 – 10,869,855 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.09 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -20.61 |
| Energy contribution | -21.80 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
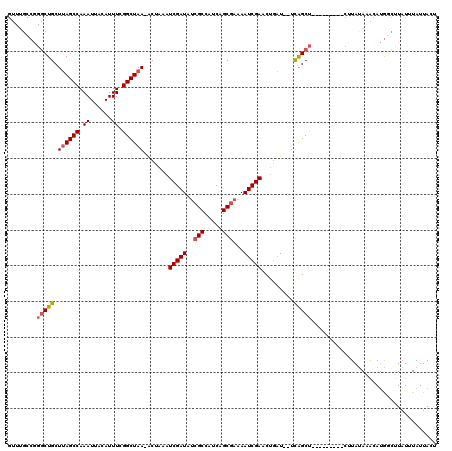
>2R_DroMel_CAF1 10869745 110 + 20766785 AGUAAUAAAUAAGCCAUGUUUGUAAG---------AGCUGAAAAUUUGUUCGAUUUUCGCUGAUGGCGAUAUCGAUUUAGU-UUAGCCGAAAUGUAAUUUGGCUAAGCAGCCCGGCAAAC .................((((((..(---------.(((..........(((((..((((.....)))).)))))....((-(((((((((......)))))))))))))))..)))))) ( -33.40) >DroSim_CAF1 10645 110 + 1 AGUAAUAAAUAAGCCAUGUUUAUAAG---------AGCUGAAAAUCAGUUCGAUUUUCGCUGAUGGCGCUAUCGAUUUAGU-UUAGCCGAAAUGUAAUUUGGCUAAGCAGCCCGGCAAAC ............(((..........(---------(((((.....))))))(((...(((.....)))..)))......((-(((((((((......))))))))))).....))).... ( -33.40) >DroEre_CAF1 11595 107 + 1 AGUAAUAAAUAAGUCAUGUGUAUAGG---------AGCCGA--AUCAGUUCGAUUUUCGCUGAUGGCGAUAUCGAUUU-GC-UUAGCCGAAAUGUAAUUUGGCUAAGCAGCCCGGCAAAC ..........................---------.((((.--......(((((..((((.....)))).))))).((-((-(((((((((......)))))))))))))..)))).... ( -36.00) >DroAna_CAF1 23077 108 + 1 AGUAAUAAAUAAAGUUUGCCUAUAUGGAGGAUGGAGGCUAA--AACAGUUCGAUUCUCGCUGAUGGCUAUAUCGAUUCAGUUUUAGCCGAAAUGUAAUUUGGCUUAGCAG---------- .............(((.(((...((...(.((...((((((--(((...(((((....((.....))...)))))....)))))))))...)).).))..)))..)))..---------- ( -24.70) >consensus AGUAAUAAAUAAGCCAUGUUUAUAAG_________AGCUGA__AUCAGUUCGAUUUUCGCUGAUGGCGAUAUCGAUUUAGU_UUAGCCGAAAUGUAAUUUGGCUAAGCAGCCCGGCAAAC ....................................((((.........(((((...(((.....)))..)))))....((.(((((((((......)))))))))))....)))).... (-20.61 = -21.80 + 1.19)

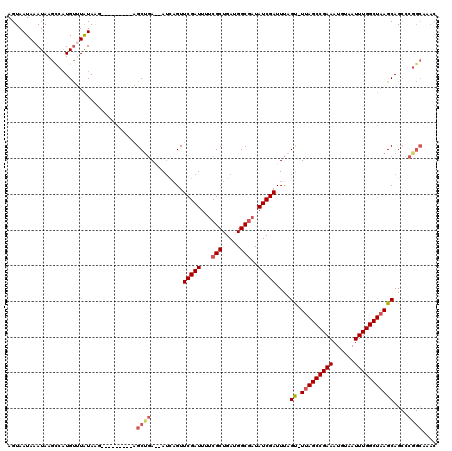
| Location | 10,869,745 – 10,869,855 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.09 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -15.45 |
| Energy contribution | -16.07 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
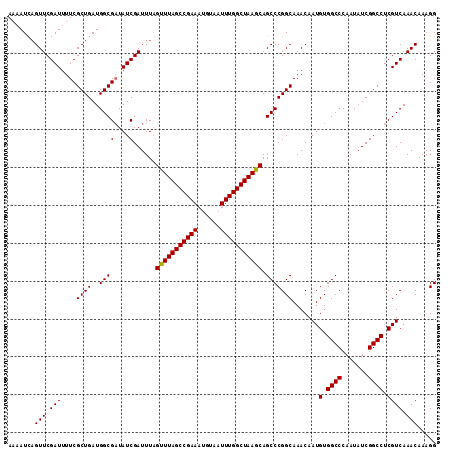
>2R_DroMel_CAF1 10869745 110 - 20766785 GUUUGCCGGGCUGCUUAGCCAAAUUACAUUUCGGCUAA-ACUAAAUCGAUAUCGCCAUCAGCGAAAAUCGAACAAAUUUUCAGCU---------CUUACAAACAUGGCUUAUUUAUUACU (((((..((((((.((((((.((......)).))))))-......(((((.((((.....))))..))))).........)))))---------)...)))))................. ( -28.10) >DroSim_CAF1 10645 110 - 1 GUUUGCCGGGCUGCUUAGCCAAAUUACAUUUCGGCUAA-ACUAAAUCGAUAGCGCCAUCAGCGAAAAUCGAACUGAUUUUCAGCU---------CUUAUAAACAUGGCUUAUUUAUUACU (((((..((((((.((((((.((......)).))))))-......(((((..(((.....)))...))))).........)))))---------)...)))))................. ( -25.90) >DroEre_CAF1 11595 107 - 1 GUUUGCCGGGCUGCUUAGCCAAAUUACAUUUCGGCUAA-GC-AAAUCGAUAUCGCCAUCAGCGAAAAUCGAACUGAU--UCGGCU---------CCUAUACACAUGACUUAUUUAUUACU ....((((((.(((((((((.((......)).))))))-))-)..(((((.((((.....))))..))))).....)--))))).---------.......................... ( -31.10) >DroAna_CAF1 23077 108 - 1 ----------CUGCUAAGCCAAAUUACAUUUCGGCUAAAACUGAAUCGAUAUAGCCAUCAGCGAGAAUCGAACUGUU--UUAGCCUCCAUCCUCCAUAUAGGCAAACUUUAUUUAUUACU ----------.......(((((((...)))).(((((((((....(((((...((.....))....)))))...)))--))))))...............)))................. ( -21.10) >consensus GUUUGCCGGGCUGCUUAGCCAAAUUACAUUUCGGCUAA_ACUAAAUCGAUAUCGCCAUCAGCGAAAAUCGAACUGAU__UCAGCU_________CUUAUAAACAUGGCUUAUUUAUUACU ........(((((.((((((.((......)).)))))).......(((((..(((.....)))...))))).........)))))................................... (-15.45 = -16.07 + 0.63)

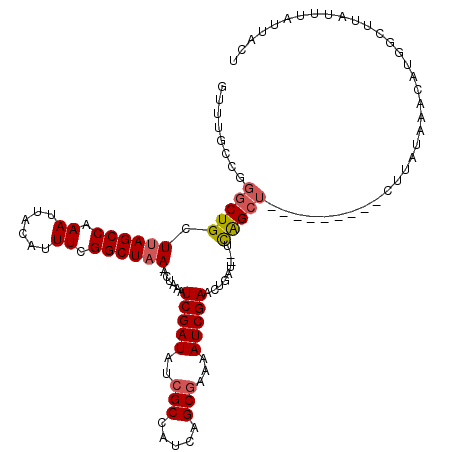
| Location | 10,869,776 – 10,869,889 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -33.45 |
| Energy contribution | -33.23 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
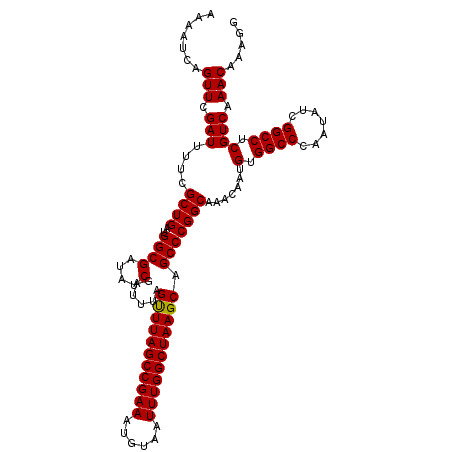
>2R_DroMel_CAF1 10869776 113 + 20766785 AAAAUUUGUUCGAUUUUCGCUGAUGGCGAUAUCGAUUUAGUUUAGCCGAAAUGUAAUUUGGCUAAGCAGCCCGGCAAACAAUGUGGCCCAAUAUCGGCCUCGUCAAACAAAGG ....((((((.(((....((((..((((....)......(((((((((((......))))))))))).))))))).......(.((((.......)))).)))).)))))).. ( -35.70) >DroSim_CAF1 10676 113 + 1 AAAAUCAGUUCGAUUUUCGCUGAUGGCGCUAUCGAUUUAGUUUAGCCGAAAUGUAAUUUGGCUAAGCAGCCCGGCAAACAAUGUGGCCCAAUAUCGGCCUCGUCAAACAAAGG .......(((.(((....((((..(((..((......))(((((((((((......))))))))))).))))))).......(.((((.......)))).)))).)))..... ( -32.00) >DroEre_CAF1 11626 110 + 1 A--AUCAGUUCGAUUUUCGCUGAUGGCGAUAUCGAUUU-GCUUAGCCGAAAUGUAAUUUGGCUAAGCAGCCCGGCAAACAAUGUGGCCCAAUCUCGGCCUCGUCAAACAAAGG .--......(((((..((((.....)))).))))).((-(((((((((((......))))))))))))).((........(((.((((.......)))).)))........)) ( -37.29) >consensus AAAAUCAGUUCGAUUUUCGCUGAUGGCGAUAUCGAUUUAGUUUAGCCGAAAUGUAAUUUGGCUAAGCAGCCCGGCAAACAAUGUGGCCCAAUAUCGGCCUCGUCAAACAAAGG .......(((.(((....((((..((((....)......(((((((((((......))))))))))).))))))).......(.((((.......)))).)))).)))..... (-33.45 = -33.23 + -0.22)

| Location | 10,869,776 – 10,869,889 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -28.89 |
| Energy contribution | -29.00 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
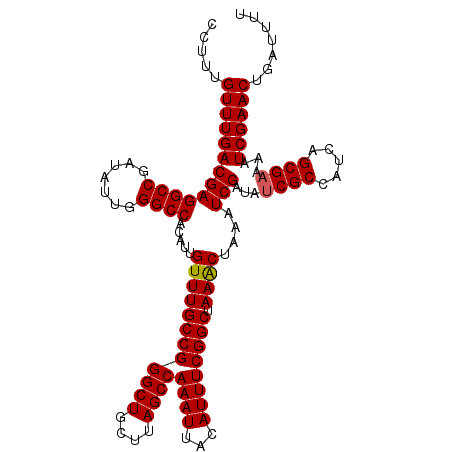
>2R_DroMel_CAF1 10869776 113 - 20766785 CCUUUGUUUGACGAGGCCGAUAUUGGGCCACAUUGUUUGCCGGGCUGCUUAGCCAAAUUACAUUUCGGCUAAACUAAAUCGAUAUCGCCAUCAGCGAAAAUCGAACAAAUUUU ..((((((((((((((((.......)))).....((((((((((((....))))((((...)))))))).))))....)))...((((.....))))...))))))))).... ( -33.30) >DroSim_CAF1 10676 113 - 1 CCUUUGUUUGACGAGGCCGAUAUUGGGCCACAUUGUUUGCCGGGCUGCUUAGCCAAAUUACAUUUCGGCUAAACUAAAUCGAUAGCGCCAUCAGCGAAAAUCGAACUGAUUUU .....(((((((((((((.......)))).....((((((((((((....))))((((...)))))))).))))....)))....(((.....)))....))))))....... ( -28.70) >DroEre_CAF1 11626 110 - 1 CCUUUGUUUGACGAGGCCGAGAUUGGGCCACAUUGUUUGCCGGGCUGCUUAGCCAAAUUACAUUUCGGCUAAGC-AAAUCGAUAUCGCCAUCAGCGAAAAUCGAACUGAU--U .....((((((...((((.......))))......(((((..((((((((((((.((......)).))))))))-)....(....))))....)))))..))))))....--. ( -34.90) >consensus CCUUUGUUUGACGAGGCCGAUAUUGGGCCACAUUGUUUGCCGGGCUGCUUAGCCAAAUUACAUUUCGGCUAAACUAAAUCGAUAUCGCCAUCAGCGAAAAUCGAACUGAUUUU .....(((((((((((((.......)))).....((((((((((((....))))((((...)))))))).))))....)))...((((.....))))...))))))....... (-28.89 = -29.00 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:27 2006