
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,851,257 – 10,851,368 |
| Length | 111 |
| Max. P | 0.951815 |

| Location | 10,851,257 – 10,851,368 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -14.93 |
| Energy contribution | -14.29 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
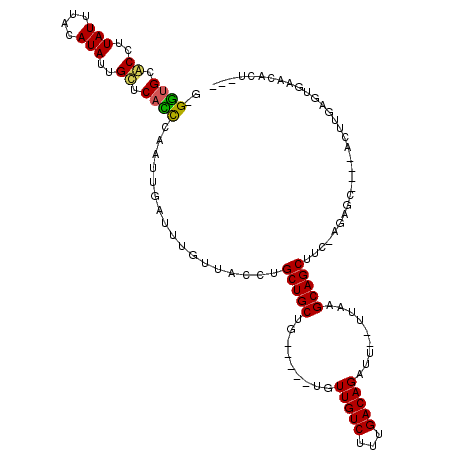
>2R_DroMel_CAF1 10851257 111 + 20766785 G-GGUGCUCCUUAUUUACAUAUUGCUCACCCAAUUGAUUUGCUACCUGCUGCUC---G-UGUUGUCUUUGACAGAUU--UUAUGCAGCUUU-GCCGCUGCCACUCCACUGAACACCAAU (-((((..(..(((....)))..)..)))))....((...((.....))...))---(-(((((((...))).....--....(((((...-...)))))..........))))).... ( -22.20) >DroVir_CAF1 19416 104 + 1 GGGUUGCACUUUAUUUACAUAUUGUUCAGUCAAUUGAUUUGUUACCUGCUGCUG-------UUGUCUUUGACAGAUUGUUUAAGCAGCUUC-GCAGC----AGUUGAAUAAACAAU--- .(..((.((..(((....)))..)).))..).((((.((((((..(((((((.(-------(((.(((.(((.....))).)))))))...-)))))----))...))))))))))--- ( -28.30) >DroPse_CAF1 3197 109 + 1 G-GGUGGGCCUUAUUUACAUAUUGCUCACCCAAUUGAUUUGUUACCUGCUGCU------UGUUGUCUUUGACAGAUU--UUGCGCAGCUUU-AGAGCUGCCACUUGAGUGAACCCUCAU (-(((((((..(((....)))..))))))))....(((((((((...((....------....))...)))))))))--.((.((((((..-..))))))))..((((......)))). ( -34.00) >DroGri_CAF1 382 105 + 1 --GGCGCACUUUAUUUACAUAUUGUUCAUUCAAUUGAUUUGUUACCUGCUGCCG---GUUGUUGUCUUUGACAGAUU-UUUAAGCAGCUUC-AGAGC----ACUUGAGUAAACAGC--- --...................(((((.((((((.((.((((....(((((....---....(((((...)))))...-....)))))...)-))).)----).)))))).))))).--- ( -21.76) >DroMoj_CAF1 16320 109 + 1 --GGUGCACUUUAUUUACAUAUUGUUCACUCAAUUGAUUUGUUACCUGCUGCUGUCUGUCGUUGUCUUUGACAGAUU-UUUAAGCAGCUUCCAUAGC----AGUUGAGUGAACAAA--- --...................(((((((((((((((...((..(.(((((...((((((((.......)))))))).-....))))).)..))...)----)))))))))))))).--- ( -41.10) >DroPer_CAF1 404 109 + 1 G-GGUGGGCCUUAUUUACAUAUUGCUCACCCAAUUGAUUUGUUACCUGCUGCU------UGUUGUCUUUGACAGAUU--UUGCGCAGCUUU-AGAGCUGCCACUUGAGUGAACCCUCAU (-(((((((..(((....)))..))))))))....(((((((((...((....------....))...)))))))))--.((.((((((..-..))))))))..((((......)))). ( -34.00) >consensus G_GGUGCACCUUAUUUACAUAUUGCUCACCCAAUUGAUUUGUUACCUGCUGCUG_____UGUUGUCUUUGACAGAUU__UUAAGCAGCUUC_AGAGC____ACUUGAGUGAACACU___ ..((((.((..(((....)))..)).)))).................(((((.........(((((...))))).........)))))............................... (-14.93 = -14.29 + -0.64)

| Location | 10,851,257 – 10,851,368 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -27.59 |
| Consensus MFE | -18.24 |
| Energy contribution | -17.55 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
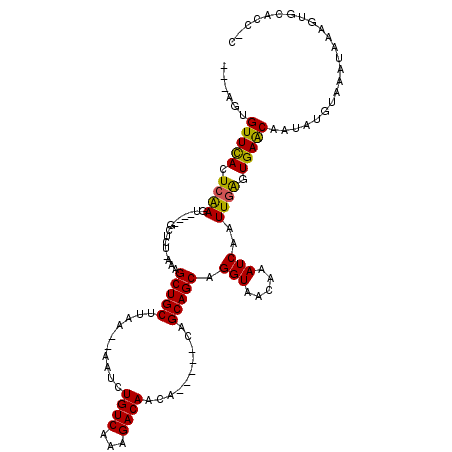
>2R_DroMel_CAF1 10851257 111 - 20766785 AUUGGUGUUCAGUGGAGUGGCAGCGGC-AAAGCUGCAUAA--AAUCUGUCAAAGACAACA-C---GAGCAGCAGGUAGCAAAUCAAUUGGGUGAGCAAUAUGUAAAUAAGGAGCACC-C .(((.(((((.(((...(((((((...-...)))))....--..(((.....))))).))-)---))))).)))..............(((((..(..(((....)))..)..))))-) ( -32.40) >DroVir_CAF1 19416 104 - 1 ---AUUGUUUAUUCAACU----GCUGC-GAAGCUGCUUAAACAAUCUGUCAAAGACAA-------CAGCAGCAGGUAACAAAUCAAUUGACUGAACAAUAUGUAAAUAAAGUGCAACCC ---((((((((.((((((----(((((-...(.(((((..((.....))..))).)).-------).)))))))(........)..)))).)))))))).((((.......)))).... ( -22.40) >DroPse_CAF1 3197 109 - 1 AUGAGGGUUCACUCAAGUGGCAGCUCU-AAAGCUGCGCAA--AAUCUGUCAAAGACAACA------AGCAGCAGGUAACAAAUCAAUUGGGUGAGCAAUAUGUAAAUAAGGCCCACC-C ....((((((((((((.((((((((..-..))))))....--.((((((...........------....)))))).......)).))))))))((..(((....)))..))..)))-) ( -29.66) >DroGri_CAF1 382 105 - 1 ---GCUGUUUACUCAAGU----GCUCU-GAAGCUGCUUAAA-AAUCUGUCAAAGACAACAAC---CGGCAGCAGGUAACAAAUCAAUUGAAUGAACAAUAUGUAAAUAAAGUGCGCC-- ---((((((((.((((..----.....-...((((((....-....((((...)))).....---.)))))).(((.....)))..)))).))))))....(((.......))))).-- ( -18.74) >DroMoj_CAF1 16320 109 - 1 ---UUUGUUCACUCAACU----GCUAUGGAAGCUGCUUAAA-AAUCUGUCAAAGACAACGACAGACAGCAGCAGGUAACAAAUCAAUUGAGUGAACAAUAUGUAAAUAAAGUGCACC-- ---.((((((((((((.(----(...((..(.(((((....-..((((((.........))))))....))))).)..))...)).))))))))))))..((((.......))))..-- ( -32.70) >DroPer_CAF1 404 109 - 1 AUGAGGGUUCACUCAAGUGGCAGCUCU-AAAGCUGCGCAA--AAUCUGUCAAAGACAACA------AGCAGCAGGUAACAAAUCAAUUGGGUGAGCAAUAUGUAAAUAAGGCCCACC-C ....((((((((((((.((((((((..-..))))))....--.((((((...........------....)))))).......)).))))))))((..(((....)))..))..)))-) ( -29.66) >consensus ___AGUGUUCACUCAAGU____GCUCU_AAAGCUGCUUAA__AAUCUGUCAAAGACAACA_____CAGCAGCAGGUAACAAAUCAAUUGAGUGAACAAUAUGUAAAUAAAGUGCACC_C ......(((((.((((...............(((((..........((((...))))..........))))).(((.....)))..)))).)))))....................... (-18.24 = -17.55 + -0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:18 2006