| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,836,968 – 10,837,087 |
| Length | 119 |
| Max. P | 0.997476 |

| Location | 10,836,968 – 10,837,087 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -23.22 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
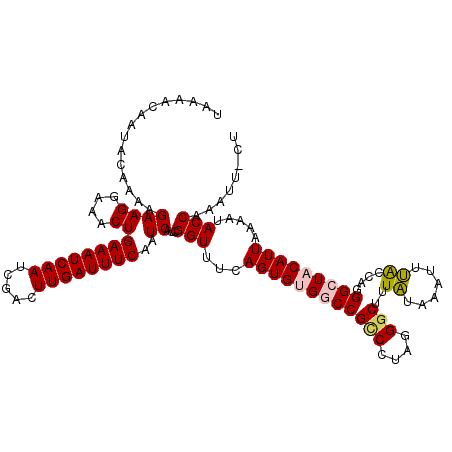
>2R_DroMel_CAF1 10836968 119 + 20766785 UAAAACAAUACAAAAGAAGGAAACUGAAAUCAAUCGACUUGAUUUCAAUCAUCGGUUUCAGUGUGGCCGCCCUAGGGCUUUGUAAGUUCACCAGGGCUACAUUAAAAUACCAAAUCACU ...............((((....))((((((((.....))))))))..))...(((...((((((((((((....)))..((......))....))))))))).....)))........ ( -30.40) >DroSec_CAF1 44330 118 + 1 UAAAACAAUACAAAAGAAGGAAACUGAAAUCAAUCGACUUGAUUUCAAUCAUCGGUUUCAGUGUGGCCGACCUAGGGCUUUAUAAUUUUAUCAGGGCUCCAUUAAAAUACCGAAUU-CU ..............(((((....)(((((((((.....)))))))))....(((((......((((((......))((((((((....))).))))).))))......))))).))-)) ( -24.80) >DroSim_CAF1 44578 118 + 1 UAAAACAAUACAAAAGAAGGAAACUGAAAUCAAUCGACUUGAUUUCAAUCAUCGGUUUCAGUGUGGCCGCCCUAGGGCUUUAUAAUUUUAUCAGGGCUCCAUUAAAAUACCGAAUU-CU ..............(((((....)(((((((((.....)))))))))....(((((...((((..((((((....)))..((......))....)))..)))).....))))).))-)) ( -29.10) >DroEre_CAF1 49970 118 + 1 UAAAACAAUACAAAAGAAGCAAACUGAAAUCAAUCGACUUGAUUUCAAUCAUCGGUUUCAGUGUGACCGUCCUGGGGCAUUAUAAAUUUACCAGGGCUACAUUAAUAUACCAAAUU-AU ...............(((((....(((((((((.....))))))))).......)))))((((((...(((((((...............))))))))))))).............-.. ( -26.46) >DroYak_CAF1 50209 118 + 1 UAAAACAAUACAAAAGAAGGAAACUGAAAUCAAUCGACUUGAUUUCAAUCAUCGGUUUGAGUGUGACCGCCCUGGGGCAUUAUAAAUUUGCUAGGGCUACAUUAAUAUACCAAAUU-AU ...............((((....))((((((((.....))))))))..))...(((...((((((...(((((..((((.........))))))))))))))).....))).....-.. ( -28.60) >consensus UAAAACAAUACAAAAGAAGGAAACUGAAAUCAAUCGACUUGAUUUCAAUCAUCGGUUUCAGUGUGGCCGCCCUAGGGCUUUAUAAAUUUACCAGGGCUACAUUAAAAUACCAAAUU_CU ...............((((....))((((((((.....))))))))..))...(((...((((((((((((....)))..((......))....))))))))).....)))........ (-23.22 = -23.62 + 0.40)


| Location | 10,836,968 – 10,837,087 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -22.76 |
| Energy contribution | -23.32 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
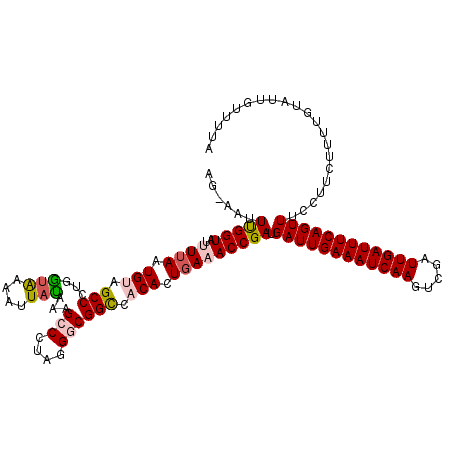
>2R_DroMel_CAF1 10836968 119 - 20766785 AGUGAUUUGGUAUUUUAAUGUAGCCCUGGUGAACUUACAAAGCCCUAGGGCGGCCACACUGAAACCGAUGAUUGAAAUCAAGUCGAUUGAUUUCAGUUUCCUUCUUUUGUAUUGUUUUA ......(((((..((((.(((.(((..((....))......(((....)))))).))).))))))))).((((((((((((.....))))))))))))..................... ( -29.60) >DroSec_CAF1 44330 118 - 1 AG-AAUUCGGUAUUUUAAUGGAGCCCUGAUAAAAUUAUAAAGCCCUAGGUCGGCCACACUGAAACCGAUGAUUGAAAUCAAGUCGAUUGAUUUCAGUUUCCUUCUUUUGUAUUGUUUUA ((-((.(((((..((((.((..(((..(((..................))))))..)).))))))))).((((((((((((.....))))))))))))...)))).............. ( -24.27) >DroSim_CAF1 44578 118 - 1 AG-AAUUCGGUAUUUUAAUGGAGCCCUGAUAAAAUUAUAAAGCCCUAGGGCGGCCACACUGAAACCGAUGAUUGAAAUCAAGUCGAUUGAUUUCAGUUUCCUUCUUUUGUAUUGUUUUA ((-((.(((((.((....(((.((((((.................))))))..)))....)).))))).((((((((((((.....))))))))))))...)))).............. ( -30.73) >DroEre_CAF1 49970 118 - 1 AU-AAUUUGGUAUAUUAAUGUAGCCCUGGUAAAUUUAUAAUGCCCCAGGACGGUCACACUGAAACCGAUGAUUGAAAUCAAGUCGAUUGAUUUCAGUUUGCUUCUUUUGUAUUGUUUUA .(-((..(((((((.....(.((((((((...((.....))...))))).((((.........))))..((((((((((((.....)))))))))))).))).)...)))))))..))) ( -25.40) >DroYak_CAF1 50209 118 - 1 AU-AAUUUGGUAUAUUAAUGUAGCCCUAGCAAAUUUAUAAUGCCCCAGGGCGGUCACACUCAAACCGAUGAUUGAAAUCAAGUCGAUUGAUUUCAGUUUCCUUCUUUUGUAUUGUUUUA .(-((..(((((((........(((((.(((.........)))...)))))(((.........)))((.((((((((((((.....))))))))))))....))...)))))))..))) ( -25.50) >consensus AG_AAUUUGGUAUUUUAAUGUAGCCCUGGUAAAAUUAUAAAGCCCUAGGGCGGCCACACUGAAACCGAUGAUUGAAAUCAAGUCGAUUGAUUUCAGUUUCCUUCUUUUGUAUUGUUUUA ......(((((..((((.(((.(((...(((....)))...(((....)))))).))).))))))))).((((((((((((.....))))))))))))..................... (-22.76 = -23.32 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:15 2006