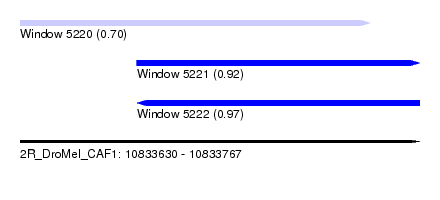
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,833,630 – 10,833,767 |
| Length | 137 |
| Max. P | 0.967198 |

| Location | 10,833,630 – 10,833,750 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.78 |
| Mean single sequence MFE | -45.77 |
| Consensus MFE | -22.40 |
| Energy contribution | -23.07 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
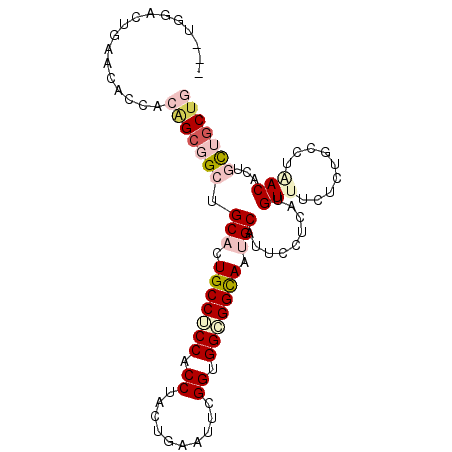
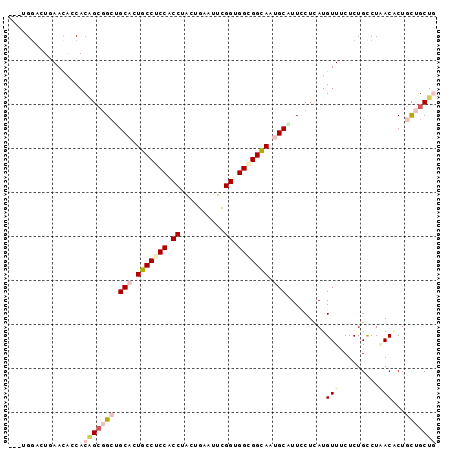
>2R_DroMel_CAF1 10833630 120 + 20766785 UGUAGUCCAUGCCUGCCUUCAUGAUCGUGUUUCGGUGCUGUAGCAAAAGUGUUAGACAGAGAAACAUGAGGAAUGCAUUACCUCCACCGAAUUCUGUAGGUGGAGGCAAUGCGGCCGCAG .........(((..((((((....(((((((((..((.(.(((((....))))).)))..))))))))).))).(((((.((((((((..........)))))))).)))))))).))). ( -47.20) >DroVir_CAF1 50110 120 + 1 UGUAGUCCAUGGCCGACUUGAUGAUUGUGUUGCGAUGCUGCAGCAACAGGGUCAGGCAGAGAAACAUAAGGAACGCAUUGCCGCCCCCAAAUUCAGCGGGUGGCGGCAGUGCAGCAGCUG (((..((((((.((..((((((..((((((((((....)))))).)))).))))))..).)...)))..)))..((((((((((((((.........))).))))))))))).))).... ( -49.30) >DroGri_CAF1 46080 120 + 1 UGUAGUCCAUCGCCGCCUUGAUAAUGGUAUUUCGAUGCUGCAGCAGCAGCGUCAGGCAGAGAAACAUAAGAAAAGCAUUGCCGCCACCAAAUUCACUAGGCGGCGGCAGUGCAGCAGCCG (((.((...((((((((........))).....((((((((....)))))))).))).))...)).........(((((((((((.((..........)).))))))))))).))).... ( -50.40) >DroWil_CAF1 63143 120 + 1 UAUAAUCCAUGCCAGCUUUCAUUAUGGUAUUACGAUGUUGCAGGAGUAGAGUCAAACAGAGAAACAUGAGAAAAGCAUUGCCGCCACCAAAUUCACUGGGUGGAGGUAAUGCAGCAGCUG ............((((((((.((((((.....)(((.((((....)))).)))...........))))))))).((((((((.(((((..........))))).))))))))....)))) ( -34.50) >DroMoj_CAF1 63510 120 + 1 UGUAGUCCAUGCCCGACUUAAUAAUGGUGUUGCGAUGCUGCAGCAGCAGUGUUAGGCAGAGGAACAUAAGGAACGCAUUGCCGCCCCCGAAUUCACCCGGUGGGGGCAGAGCAGCCGCUG (((..(((.((((..((.......((.(((((((....))))))).))..))..))))..)))..))).((...((.(((((.((((((........))).)))))))).))..)).... ( -46.20) >DroAna_CAF1 40820 120 + 1 UGUAGUCCAUGGCCCCCUUGAUGAUGGUGUUCCUGUGCUGCAGCAGCAGGGUGAGGCAGAGGAACAUGAGGAAGGCAUUACCGCCACCGAACUCAGACGGCGGAGGCAGAGCCGCCGCCG ((.((((..((((..((((.......((((((((.((((.((.(.....).)).)))).))))))))))))..((.....))))))..).)))))..((((((.(((...))).)))))) ( -47.01) >consensus UGUAGUCCAUGCCCGCCUUGAUGAUGGUGUUGCGAUGCUGCAGCAGCAGGGUCAGGCAGAGAAACAUAAGGAAAGCAUUGCCGCCACCAAAUUCACUAGGUGGAGGCAGUGCAGCAGCUG ..........................(((((.(..((((...((......))..))))..).))))).......((((((((.((.((..........)).)).))))))))........ (-22.40 = -23.07 + 0.67)

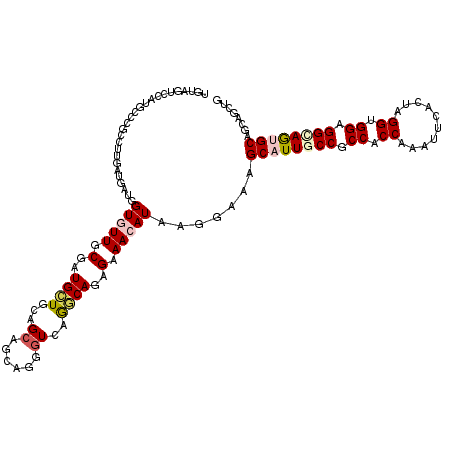
| Location | 10,833,670 – 10,833,767 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -17.24 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10833670 97 + 20766785 UAGCAAAAGUGUUAGACAGAGAAACAUGAGGAAUGCAUUACCUCCACCGAAUUCUGUAGGUGGAGGCAAUGCGGCCGCAGUGGUAUUCACUGCACUG (((((....)))))..(((..........((..((((((.((((((((..........)))))))).)))))).))((((((.....)))))).))) ( -37.60) >DroVir_CAF1 50150 94 + 1 CAGCAACAGGGUCAGGCAGAGAAACAUAAGGAACGCAUUGCCGCCCCCAAAUUCAGCGGGUGGCGGCAGUGCAGCAGCUGUGCUAUUGAUCCCG--- ........((((((((((.((...(....)....((((((((((((((.........))).))))))))))).....)).))))..))))))..--- ( -41.00) >DroGri_CAF1 46120 94 + 1 CAGCAGCAGCGUCAGGCAGAGAAACAUAAGAAAAGCAUUGCCGCCACCAAAUUCACUAGGCGGCGGCAGUGCAGCAGCCGUGCUGUUCAGGCCG--- ..((.((((((...(((.......(....)....(((((((((((.((..........)).)))))))))))....))).))))))....))..--- ( -38.20) >DroEre_CAF1 46667 97 + 1 UAGUAGAAGUGUGAGACAGAGAAACAUGAGGAAUGCAUUACCUCCGCCGAAUUCUGUAGGUGGAGGCAAUGCAGCCGCUGUGGUGUUCAGUCCAGUG ...........((.(((.(((...((((.((..((((((.((((((((..........)))))))).)))))).))..))))...))).)))))... ( -33.20) >DroMoj_CAF1 63550 94 + 1 CAGCAGCAGUGUUAGGCAGAGGAACAUAAGGAACGCAUUGCCGCCCCCGAAUUCACCCGGUGGGGGCAGAGCAGCCGCUGUGCUAUUCAGCCCA--- ..(((((.(((((.........)))))..((...((.(((((.((((((........))).)))))))).))..)))))))(((....)))...--- ( -33.20) >DroAna_CAF1 40860 94 + 1 CAGCAGCAGGGUGAGGCAGAGGAACAUGAGGAAGGCAUUACCGCCACCGAACUCAGACGGCGGAGGCAGAGCCGCCGCCGUGGCGUUUACUCC---G ........(((((((((.(((........((..(((......))).))...)))..(((((((.(((...))).))))))).)).))))).))---. ( -34.60) >consensus CAGCAGCAGUGUCAGGCAGAGAAACAUAAGGAACGCAUUACCGCCACCGAAUUCAGUAGGUGGAGGCAGUGCAGCCGCUGUGCUAUUCAGUCCA___ ..................(((...(((..((...(((((.(((((.((..........)).))))).)))))..))...)))...)))......... (-17.24 = -18.02 + 0.78)


| Location | 10,833,670 – 10,833,767 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -20.24 |
| Energy contribution | -21.60 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10833670 97 - 20766785 CAGUGCAGUGAAUACCACUGCGGCCGCAUUGCCUCCACCUACAGAAUUCGGUGGAGGUAAUGCAUUCCUCAUGUUUCUCUGUCUAACACUUUUGCUA ....((((((.....))))))(((.((((((((((((((..........))))))))))))))........((((.........)))).....))). ( -38.20) >DroVir_CAF1 50150 94 - 1 ---CGGGAUCAAUAGCACAGCUGCUGCACUGCCGCCACCCGCUGAAUUUGGGGGCGGCAAUGCGUUCCUUAUGUUUCUCUGCCUGACCCUGUUGCUG ---.(((.(((...(((.((..((((((.(((((((.((((.......))))))))))).))))........))..)).))).))))))........ ( -34.60) >DroGri_CAF1 46120 94 - 1 ---CGGCCUGAACAGCACGGCUGCUGCACUGCCGCCGCCUAGUGAAUUUGGUGGCGGCAAUGCUUUUCUUAUGUUUCUCUGCCUGACGCUGCUGCUG ---.........(((((((((....(((.((((((((((..........)))))))))).))).........(((.........))))))).))))) ( -37.60) >DroEre_CAF1 46667 97 - 1 CACUGGACUGAACACCACAGCGGCUGCAUUGCCUCCACCUACAGAAUUCGGCGGAGGUAAUGCAUUCCUCAUGUUUCUCUGUCUCACACUUCUACUA ....((((.(((....((((.((.((((((((((((.((..........)).))))))))))))..)).).))))))...))))............. ( -29.60) >DroMoj_CAF1 63550 94 - 1 ---UGGGCUGAAUAGCACAGCGGCUGCUCUGCCCCCACCGGGUGAAUUCGGGGGCGGCAAUGCGUUCCUUAUGUUCCUCUGCCUAACACUGCUGCUG ---...(((....))).(((((((.((..(((((((.(((((....)))))))).))))..)).....(((.((......)).)))....))))))) ( -33.40) >DroAna_CAF1 40860 94 - 1 C---GGAGUAAACGCCACGGCGGCGGCUCUGCCUCCGCCGUCUGAGUUCGGUGGCGGUAAUGCCUUCCUCAUGUUCCUCUGCCUCACCCUGCUGCUG .---((.(....).)).((((((((((..((((.((((((........)))))).))))..)))........((......))........))))))) ( -34.60) >consensus ___UGGACUGAACACCACAGCGGCUGCACUGCCUCCACCUACUGAAUUCGGUGGCGGCAAUGCAUUCCUCAUGUUUCUCUGCCUAACACUGCUGCUG .................(((((((.(((.(((((((.((..........)).))))))).))).........(((.........)))...))))))) (-20.24 = -21.60 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:12 2006