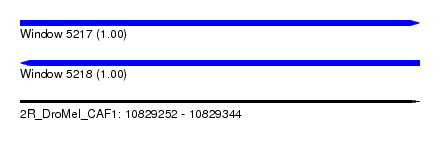
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,829,252 – 10,829,344 |
| Length | 92 |
| Max. P | 0.999984 |

| Location | 10,829,252 – 10,829,344 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 64.69 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -12.14 |
| Energy contribution | -13.70 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.54 |
| SVM decision value | 5.16 |
| SVM RNA-class probability | 0.999976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
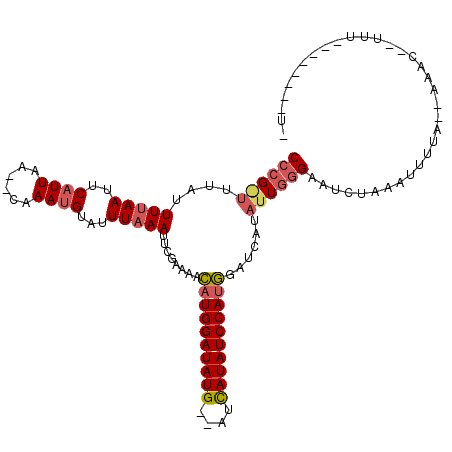
>2R_DroMel_CAF1 10829252 92 + 20766785 CCCGGUUUAUUUUAAUUCAUUUA--CAAAUGUAUUUAAAUUCAAAAUCAUGGAUAUG--AUCAUAUCCAUGGUUCAUAUUGGGAAUCUCAAUUUUC--------------------- ((((((....(((((..((((..--..))))...))))).....((((((((((((.--...))))))))))))...)))))).............--------------------- ( -21.50) >DroSim_CAF1 36838 104 + 1 CCCGAUUUAUUUUAAUUCAUUAA--CAAAUGUUUUUAAAUUGGAAAACAUGGAUAUG--AUCAUAUCCAUGGACCAUAUUGGGAAUCUACAUUUUUGUAAACUUUUU--------U- ((((((...((((((((.(..((--(....)))..).))))))))..(((((((((.--...)))))))))......))))))....((((....))))........--------.- ( -24.10) >DroEre_CAF1 42138 113 + 1 CC-G--UUUUUUUAAGUCAUUAGAGGAAACGUAUUUUAAUUUGAUAAUGUGGAUAUGUGGAUAUAUCCAAAGAUCAUGUUGUGAAUCGUAAUUCUA-AAAACAAUUUGAAUUCUAUA ..-.--.........((..((((((...(((.((((((((.((((....((((((((....))))))))...)))).)))).)))))))..)))))-)..))............... ( -22.00) >consensus CCCG_UUUAUUUUAAUUCAUUAA__CAAAUGUAUUUAAAUUCGAAAACAUGGAUAUG__AUCAUAUCCAUGGAUCAUAUUGGGAAUCUAAAUUUUA__AAAC__UUU________U_ ((((((....(((((..((((......))))...)))))........((((((((((....))))))))))......)))))).................................. (-12.14 = -13.70 + 1.56)

| Location | 10,829,252 – 10,829,344 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 64.69 |
| Mean single sequence MFE | -19.18 |
| Consensus MFE | -11.73 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.61 |
| SVM decision value | 5.34 |
| SVM RNA-class probability | 0.999984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
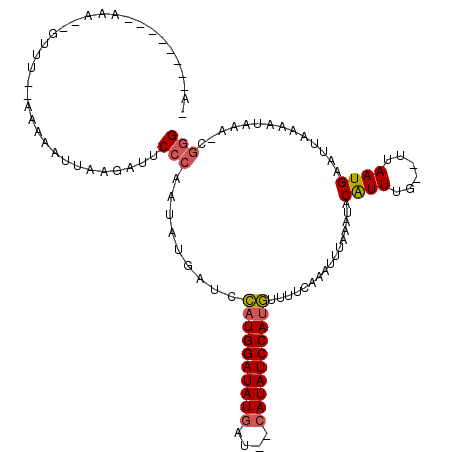
>2R_DroMel_CAF1 10829252 92 - 20766785 ---------------------GAAAAUUGAGAUUCCCAAUAUGAACCAUGGAUAUGAU--CAUAUCCAUGAUUUUGAAUUUAAAUACAUUUG--UAAAUGAAUUAAAAUAAACCGGG ---------------------.............(((.........(((((((((...--.)))))))))(((((((.((((..(((....)--))..))))))))))).....))) ( -19.70) >DroSim_CAF1 36838 104 - 1 -A--------AAAAAGUUUACAAAAAUGUAGAUUCCCAAUAUGGUCCAUGGAUAUGAU--CAUAUCCAUGUUUUCCAAUUUAAAAACAUUUG--UUAAUGAAUUAAAAUAAAUCGGG -.--------....((((((((....))))))))(((.....((..(((((((((...--.)))))))))....))((((((..(((....)--))..))))))..........))) ( -23.20) >DroEre_CAF1 42138 113 - 1 UAUAGAAUUCAAAUUGUUUU-UAGAAUUACGAUUCACAACAUGAUCUUUGGAUAUAUCCACAUAUCCACAUUAUCAAAUUAAAAUACGUUUCCUCUAAUGACUUAAAAAAA--C-GG ...............(((.(-((((...(((..........((((...(((((((......)))))))....))))..........)))....))))).))).........--.-.. ( -14.65) >consensus _A________AAA__GUUU__AAAAAUUAAGAUUCCCAAUAUGAUCCAUGGAUAUGAU__CAUAUCCAUGUUUUCAAAUUUAAAUACAUUUG__UUAAUGAAUUAAAAUAAA_CGGG ..................................(((.........((((((((((....))))))))))................((((......))))..............))) (-11.73 = -12.40 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:08 2006