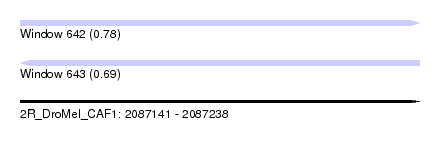
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,087,141 – 2,087,238 |
| Length | 97 |
| Max. P | 0.777359 |

| Location | 2,087,141 – 2,087,238 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -21.11 |
| Energy contribution | -22.70 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
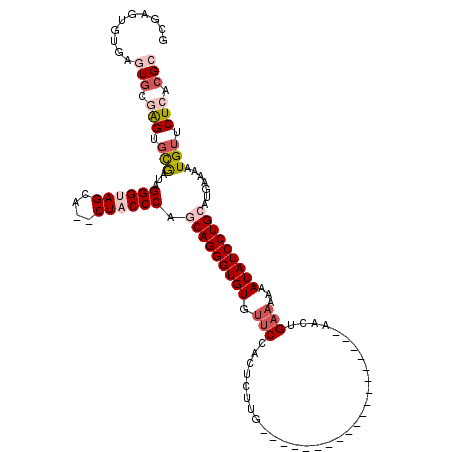
>2R_DroMel_CAF1 2087141 97 + 20766785 GCAAGUGUGAGUACGAGUGCGAUAGGGUAGCA--CUACCCAGCAGGGUGUGUUCCACUCUUG-------------------AACUGAAAAAAUAUCCUGCAUGAAAAUGUUCUCACGC ....(((((((.(((.........(((((...--.))))).(((((((((.(((........-------------------....)))...))))))))).......))).))))))) ( -32.60) >DroSim_CAF1 74214 97 + 1 GCAAGUGUGCGUGCGAGUGCGAUAGGGUAGCA--CUACCCAGCAGGGUGUGUUCCACUCUUG-------------------AACUGAAAAAAUAUCCUGCAUGAAAAUGUUCUCACGC (((....)))(((.(((.(((...(((((...--.))))).(((((((((.(((........-------------------....)))...))))))))).......))).))).))) ( -31.00) >DroEre_CAF1 69684 97 + 1 GCGAGUGUGAGUGCGAGUGCGAUAGGGUAGCA--CUACCCAGCAGGGUGUGUUCCACUCUUG-------------------AACGGAAAAAAUAUCCUGCAUGAAAAUGUUCUCACGC ....(((((((.(((.........(((((...--.))))).(((((((((.((((.......-------------------...))))...))))))))).......))).))))))) ( -37.10) >DroYak_CAF1 73710 97 + 1 GAUUAUUCGAGUGUGAGUGCAAUAGGGUAGCA--CUACCCAGCAGGGUGUGUUCCAUUCUUG-------------------AACUGAAAAAAUAUCCUGCAUGAAAAUGUUCUCACGG ...........((((((.(((...(((((...--.))))).(((((((((.(((..((....-------------------))..)))...))))))))).......))).)))))). ( -32.00) >DroMoj_CAF1 122036 105 + 1 GUGCGUGCUUGUGUUGGUCUGAUAGGGUAGCAGACUUCCCACCAGGGUGUGUUCAACUCUUGAUGAGC-------------AGCCGCUAAAAUAUCCUGCAUGAAAAUGUUCUCACGC ..(((((((.(((..((((((.........))))))...))).))(((.((((((.(....).)))))-------------)))).............((((....))))...))))) ( -28.00) >DroAna_CAF1 73906 113 + 1 GUGAGUG----UGUCAGUGCGAUGGGGUAGCA-ACUUCCCACCAGGGUGUGUUCCACACUUGAUGAGCAGCCACCGCAGCGAACUGAAAAAAUAUCCUGCAUGAAAAUGUUCUCACGC (((((..----..(((((.((.((.(((.((.-.(((..((....((((((...))))))))..)))..)).))).)).)).)))))...........((((....)))).))))).. ( -33.80) >consensus GCGAGUGUGAGUGCGAGUGCGAUAGGGUAGCA__CUACCCAGCAGGGUGUGUUCCACUCUUG___________________AACUGAAAAAAUAUCCUGCAUGAAAAUGUUCUCACGC ..........(((.(((.(((...((((((....)))))).(((((((((.(((...............................)))...))))))))).......))).))).))) (-21.11 = -22.70 + 1.59)

| Location | 2,087,141 – 2,087,238 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
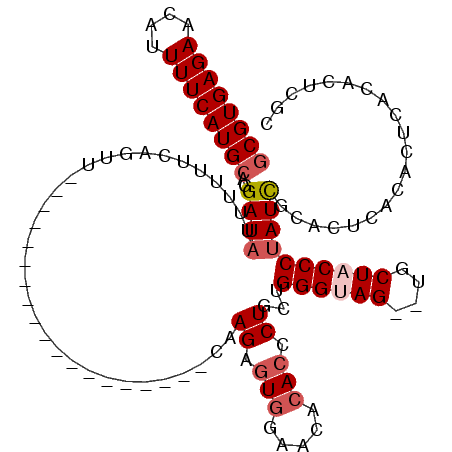
>2R_DroMel_CAF1 2087141 97 - 20766785 GCGUGAGAACAUUUUCAUGCAGGAUAUUUUUUCAGUU-------------------CAAGAGUGGAACACACCCUGCUGGGUAG--UGCUACCCUAUCGCACUCGUACUCACACUUGC ((((((((....)))))))).(((......)))....-------------------((((.((((.((..((((....))))((--(((.........))))).)).).))).)))). ( -27.20) >DroSim_CAF1 74214 97 - 1 GCGUGAGAACAUUUUCAUGCAGGAUAUUUUUUCAGUU-------------------CAAGAGUGGAACACACCCUGCUGGGUAG--UGCUACCCUAUCGCACUCGCACGCACACUUGC (((((.((..........(((((......(((((.((-------------------...)).))))).....))))).(((((.--...)))))........)).)))))........ ( -26.60) >DroEre_CAF1 69684 97 - 1 GCGUGAGAACAUUUUCAUGCAGGAUAUUUUUUCCGUU-------------------CAAGAGUGGAACACACCCUGCUGGGUAG--UGCUACCCUAUCGCACUCGCACUCACACUCGC ((((((((....)))))))).(((.......)))...-------------------...(((((((....((((....))))((--(((.........))))).....)).))))).. ( -30.50) >DroYak_CAF1 73710 97 - 1 CCGUGAGAACAUUUUCAUGCAGGAUAUUUUUUCAGUU-------------------CAAGAAUGGAACACACCCUGCUGGGUAG--UGCUACCCUAUUGCACUCACACUCGAAUAAUC ..(((((...........(((((......(((((...-------------------......))))).....))))).(((((.--...))))).......)))))............ ( -23.50) >DroMoj_CAF1 122036 105 - 1 GCGUGAGAACAUUUUCAUGCAGGAUAUUUUAGCGGCU-------------GCUCAUCAAGAGUUGAACACACCCUGGUGGGAAGUCUGCUACCCUAUCAGACCAACACAAGCACGCAC ((((((((....))))))))...........((((((-------------((((.....))))..........((((((((.((....)).))).))))).........))).))).. ( -31.30) >DroAna_CAF1 73906 113 - 1 GCGUGAGAACAUUUUCAUGCAGGAUAUUUUUUCAGUUCGCUGCGGUGGCUGCUCAUCAAGUGUGGAACACACCCUGGUGGGAAGU-UGCUACCCCAUCGCACUGACA----CACUCAC ((((((((....))))))))...........(((((.((.((.((((((..(((((((.(((((...)))))..)))))))....-.)))))).)).)).)))))..----....... ( -41.00) >consensus GCGUGAGAACAUUUUCAUGCAGGAUAUUUUUUCAGUU___________________CAAGAGUGGAACACACCCUGCUGGGUAG__UGCUACCCUAUCGCACUCACACUCACACUCGC ((((((((....))))))))..((((................................((.(((.....))).))...((((((....)))))))))).................... (-18.02 = -18.88 + 0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:17 2006