| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,819,972 – 10,820,063 |
| Length | 91 |
| Max. P | 0.825772 |

| Location | 10,819,972 – 10,820,063 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -19.52 |
| Energy contribution | -21.04 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10819972 91 + 20766785 CGGCUGUCAUAAUUUUCAAUUUUAUACCUCCCGAGCUUUAUGCUCACCUAUCCGGGAGGAGUCCGGGCUAUGGACCCCGGAGUUUAUGCCG ((((......................((((((((((.....))))........))))))..((((((........))))))......)))) ( -31.60) >DroSec_CAF1 27348 91 + 1 CGGCUGUCAUAAUUUUCAAUUUUAUACCUCCCGAGCUUUAUGCUCACCUAACCGGGAGGAGUCCGGACUAUGGACUCCGGAGUUUAUGCCG ((((....((((.........))))..((((.((((.....)))).(((....))).((((((((.....)))))))))))).....)))) ( -31.30) >DroSim_CAF1 27613 91 + 1 CGGCUGUCAUAAUUUUCAAUUUUAUACCUCCCGAGCUUUAUGCUCACCUAACCGAGAGGAGUCCGGACUAUGGACUCCGGAGUUUAUGCCG ((((....((((.........))))..((((.((((.....))))............((((((((.....)))))))))))).....)))) ( -30.00) >DroEre_CAF1 32253 88 + 1 CGUCUGUCAUAAUUUUCAAUUUUAUACCUCCUGAGCUUUAUGCUCACCUACCCGGG---AGUCCGGACUAUGGACUCCGGAGUUUAUGCCG .....(.(((((.((((..............(((((.....)))))........((---((((((.....)))))))))))).))))).). ( -26.40) >DroYak_CAF1 33425 84 + 1 CGUCUGUCAUAAUUUUCAAUUUUAUACCUCCCGAGCUUUAUGCUCACCUAUCCGGG---AGUACGGACUA----CUCCGAAGUUUAUGCCG .....(.(((((.((((...............((((.....)))).........((---((((.....))----)))))))).))))).). ( -20.20) >consensus CGGCUGUCAUAAUUUUCAAUUUUAUACCUCCCGAGCUUUAUGCUCACCUAACCGGGAGGAGUCCGGACUAUGGACUCCGGAGUUUAUGCCG ((((......................((((((((((.....))))........))))))..((((((........))))))......)))) (-19.52 = -21.04 + 1.52)

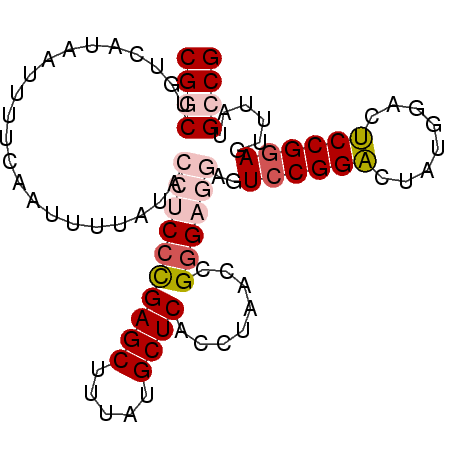
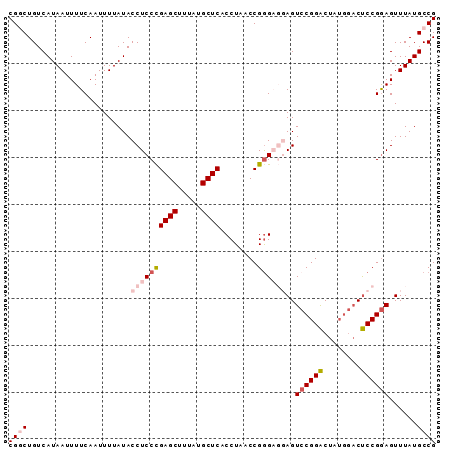
| Location | 10,819,972 – 10,820,063 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -20.82 |
| Energy contribution | -21.98 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10819972 91 - 20766785 CGGCAUAAACUCCGGGGUCCAUAGCCCGGACUCCUCCCGGAUAGGUGAGCAUAAAGCUCGGGAGGUAUAAAAUUGAAAAUUAUGACAGCCG ((((......((((((........))))))..((((((........((((.....))))))))))......................)))) ( -31.20) >DroSec_CAF1 27348 91 - 1 CGGCAUAAACUCCGGAGUCCAUAGUCCGGACUCCUCCCGGUUAGGUGAGCAUAAAGCUCGGGAGGUAUAAAAUUGAAAAUUAUGACAGCCG ((((.....(((((((((((.......)))))))..((.....))(((((.....))))))))).(((((.........)))))...)))) ( -30.70) >DroSim_CAF1 27613 91 - 1 CGGCAUAAACUCCGGAGUCCAUAGUCCGGACUCCUCUCGGUUAGGUGAGCAUAAAGCUCGGGAGGUAUAAAAUUGAAAAUUAUGACAGCCG ((((.....(((((((((((.......)))))))...........(((((.....))))))))).(((((.........)))))...)))) ( -30.30) >DroEre_CAF1 32253 88 - 1 CGGCAUAAACUCCGGAGUCCAUAGUCCGGACU---CCCGGGUAGGUGAGCAUAAAGCUCAGGAGGUAUAAAAUUGAAAAUUAUGACAGACG .(.(((((((((.(((((((.......)))))---)).))))...(((((.....)))))...................))))).)..... ( -26.60) >DroYak_CAF1 33425 84 - 1 CGGCAUAAACUUCGGAG----UAGUCCGUACU---CCCGGAUAGGUGAGCAUAAAGCUCGGGAGGUAUAAAAUUGAAAAUUAUGACAGACG .(.(((((..((((((.----...)))...((---(((........((((.....)))))))))..........)))..))))).)..... ( -19.10) >consensus CGGCAUAAACUCCGGAGUCCAUAGUCCGGACUCCUCCCGGAUAGGUGAGCAUAAAGCUCGGGAGGUAUAAAAUUGAAAAUUAUGACAGCCG ((((.....(((((((((((.......)))))))..((.....))(((((.....))))))))).(((((.........)))))...)))) (-20.82 = -21.98 + 1.16)


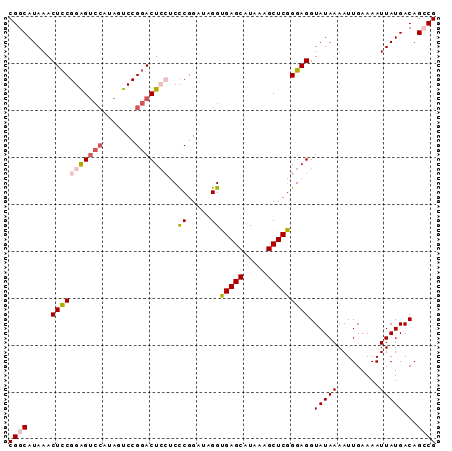
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:04 2006