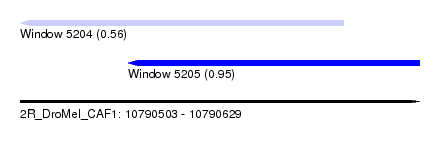
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,790,503 – 10,790,629 |
| Length | 126 |
| Max. P | 0.948889 |

| Location | 10,790,503 – 10,790,605 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -7.46 |
| Energy contribution | -9.60 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.25 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
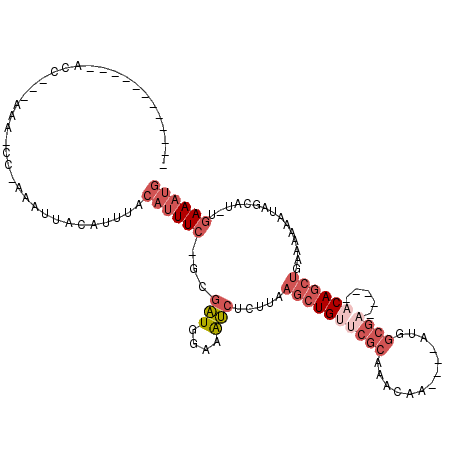
>2R_DroMel_CAF1 10790503 102 - 20766785 UUC-GCGAUGGAAAUCUCUUGAGCUGCUCGCAAACAA----AUGGCGA-----ACAGCUGAAAAAA-AGCAU-UGAAAUGGAGCUGUU--UUCAUUGUUAUUCAAG-CAUCCCUGCA- ...-((((((((((((((((.(((((.((((......----...))))-----.))))).))....-..(((-....))))))..)))--)))))))).......(-((....))).- ( -27.10) >DroPse_CAF1 1029 99 - 1 UUC-AGCCUGGAAGGUACUCGACCUGCCAGCAAACAAUGAAAUGG-GU-----ACACAU-----AAUAGAAUGUGAAUUGGAGG-GCU--UUCAUUAUCCGUUUGGUUGUACCC---- ...-.((.(((.((((.....)))).)))))..(((((.((((((-((-----((((((-----......)))))...(((((.-..)--)))).))))))))).)))))....---- ( -29.50) >DroSec_CAF1 1100 103 - 1 UUC-GCGAUGGAAAUCUCUUGAGCUGUUCGCAAACAA----AUGGCGA-----ACAGCUGAAAAAAUAGCAU-UGAAAUGGAGCCGUU--UUCAUUGUUAUUCAAG-CAUCCCUGCA- ...-(((((....))).((((((((((((((......----...))))-----)))))).....(((((((.-(((((((....)).)--)))).)))))))))))-.......)).- ( -33.40) >DroEre_CAF1 1100 103 - 1 UUC-GCGAUGGAAAUCUCUUAAGCUGUUCGCAAACAA----AUGGCGA-----ACAGCUGAAAAAAUAGCAU-CGAAAUGGAGCCGUU--UUCAUUGUUAUUCAAG-CAUUCCUCCA- ...-(.((.((((........((((((((((......----...))))-----)))))).....(((((((.-.((((((....))))--))...)))))))....-..))))))).- ( -29.90) >DroYak_CAF1 1634 105 - 1 UUC-GCGAUGGAAAUCCCUUAAGCUGUUCGCAAACAA----AUGGCGA-----ACAGCUGAAAAAAUAGCAU-CGAAACGGAGCCGUUUUUUCAUUGUUAUUCAAG-CAUCCCUGCA- ...-(((..(((.........((((((((((......----...))))-----))))))(((((((..((..-((...))..))..))))))).............-..))).))).- ( -31.70) >DroAna_CAF1 1102 108 - 1 UUAGGCGAUGAAAAUCUCUUAAGCUGUCCGCAAACAA----UGGCCGACAAGCACAGCA-AAAAAAUUGAAU-UGAAAUGGAGGCGUU---UCAUUGUUAUUCAAG-UCCCGCUGCAG (((((.(((....))).)))))((((((.((......----..)).))).))).((((.-......((((((-(((((((....))))---))).....)))))).-....))))... ( -27.60) >consensus UUC_GCGAUGGAAAUCUCUUAAGCUGUUCGCAAACAA____AUGGCGA_____ACAGCUGAAAAAAUAGCAU_UGAAAUGGAGCCGUU__UUCAUUGUUAUUCAAG_CAUCCCUGCA_ ....((((((((.........((((((((((.............)))).....)))))).................((((....))))..)))))))).................... ( -7.46 = -9.60 + 2.14)



| Location | 10,790,537 – 10,790,629 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.44 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -5.84 |
| Energy contribution | -7.92 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.27 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10790537 92 - 20766785 ------------ACC---AAAACC-AAAUUACAUUUACAUUUC-GCGAUGGAAAUCUCUUGAGCUGCUCGCAAACAA----AUGGCGA-----ACAGCUGAAAAAA-AGCAU-UGAAAUG ------------...---......-............((((((-(((((....)))..((.(((((.((((......----...))))-----.))))).))....-.))..-.)))))) ( -21.20) >DroSec_CAF1 1134 92 - 1 ------------ACC---AAA-CC-CAAUUACAUUUACAUUUC-GCGAUGGAAAUCUCUUGAGCUGUUCGCAAACAA----AUGGCGA-----ACAGCUGAAAAAAUAGCAU-UGAAAUG ------------...---...-..-............((((((-(((((....)))..((.((((((((((......----...))))-----)))))).))......))..-.)))))) ( -25.40) >DroEre_CAF1 1134 92 - 1 ------------ACC---AAA-CC-AAAUUACAUUUACAUUUC-GCGAUGGAAAUCUCUUAAGCUGUUCGCAAACAA----AUGGCGA-----ACAGCUGAAAAAAUAGCAU-CGAAAUG ------------...---...-..-............((((((-(.(((....))).....((((((((((......----...))))-----)))))).............-))))))) ( -25.00) >DroYak_CAF1 1670 92 - 1 ------------ACC---AAA-CC-AAAUUACAUUUACAUUUC-GCGAUGGAAAUCCCUUAAGCUGUUCGCAAACAA----AUGGCGA-----ACAGCUGAAAAAAUAGCAU-CGAAACG ------------...---...-..-..................-.(((((...........((((((((((......----...))))-----))))))..........)))-))..... ( -21.90) >DroAna_CAF1 1136 109 - 1 CUUUCAAAGAAAAAC---AAA-CA-AAAUUACAUUUACAUUUAGGCGAUGAAAAUCUCUUAAGCUGUCCGCAAACAA----UGGCCGACAAGCACAGCA-AAAAAAUUGAAU-UGAAAUG .((((((........---...-..-...............(((((.(((....))).)))))((((((.((......----..)).))).)))......-...........)-))))).. ( -18.70) >DroPer_CAF1 1060 95 - 1 ------------ACAAGAAAA-CCAAAAUUAUAUUCAAAUUUC-AGCCUGGAAGGUACUCGACCUGCCAGCAAACAAUGAAAUGG-GU-----ACACAU-----AAUAGAAUGUGAAUUG ------------.........-.........((((((..((((-(..((((.((((.....)))).)))).......))))))))-))-----)(((((-----......)))))..... ( -18.30) >consensus ____________ACC___AAA_CC_AAAUUACAUUUACAUUUC_GCGAUGGAAAUCUCUUAAGCUGUUCGCAAACAA____AUGGCGA_____ACAGCUGAAAAAAUAGCAU_UGAAAUG .....................................((((((...(((....))).....((((((((((.............)))).....))))))...............)))))) ( -5.84 = -7.92 + 2.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:55 2006