| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,765,323 – 10,765,428 |
| Length | 105 |
| Max. P | 0.963953 |

| Location | 10,765,323 – 10,765,428 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.71 |
| Mean single sequence MFE | -20.61 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.78 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
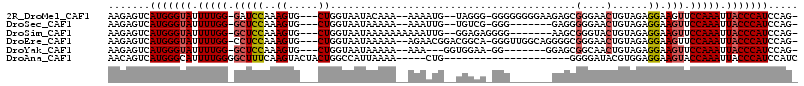
>2R_DroMel_CAF1 10765323 105 + 20766785 -CUGGAUGGGUAAUUUGGAACUUCCUCUACAGUUCCCGCUCUUCCCCCCCC-CCCUA--CAUUUU--UUUGUAUUACCAG---CACUUUGGAUC-CCAAAAUACCCAUGACUCUU -....(((((((.(((((.................................-...((--((....--..))))...((((---....))))...-))))).)))))))....... ( -19.90) >DroSec_CAF1 1086 98 + 1 -CUGGAUGGGUAAUUUGGAACUUCCUCUACAGUUCCCCCUC-------CCC-CGACA--CAAUUU--UUUUUAUUACCAG---CACUUUGGAGC-CCAAAAUACCCAUGACUCUU -....(((((((.(((((((((........)))))).....-------...-.....--......--.........((((---....))))...-..))).)))))))....... ( -18.30) >DroSim_CAF1 1164 101 + 1 -CUGGAUGGGUAAUUUGGAACUUCCUCUACAGUACCCGCUU-------CCCCUCUCC--CAAUUUUUUUUUUAUUACCAG---CACUUUGGAGC-CCAAAAUACCCAUGACUCUU -....(((((((.(((((......(((((.(((....(((.-------.........--...................))---)))).))))).-))))).)))))))....... ( -18.75) >DroEre_CAF1 1120 107 + 1 -CUGGAUGGGUAAUUUGGAACUUCCUCUACAGUUCCCGCCCCUGCCAACCC-UGCCGUCCGUUCU--UUUUUAUUACCAG---CACUUUGGAGG-CCAAAAUACCCAUGACUCUU -....(((((((.(((((((((........)))))).(((....((((...-(((..........--............)---))..)))).))-).))).)))))))....... ( -23.45) >DroYak_CAF1 1094 97 + 1 -CUGGAUGGGUAAUUUGGAACUUCCUCUACAGUUGCCGCUCC-------CC-UUCCACC---UUU--UUUUUAUUACCAG---CACUUUGGAGC-CCAAAAUACCCAUGACUCUU -....(((((((.(((((((((........))))...(((((-------..-.......---...--.............---......)))))-))))).)))))))....... ( -21.88) >DroAna_CAF1 14867 89 + 1 GAUGGAUGGGUAAUUUGGUACUUCCUCCACGUAUCCCC---------------------CAG-----UUUUAAUGGCCAGUAGUACUUGAAAGCCCCAAAAUGCCCAUGACUGUU ((((((((((((.(((((..(((....((.((((..((---------------------((.-----......)))...)..)))).)).)))..))))).)))))))..))))) ( -21.40) >consensus _CUGGAUGGGUAAUUUGGAACUUCCUCUACAGUUCCCGCUC_______CCC_UCCCA__CAAUUU__UUUUUAUUACCAG___CACUUUGGAGC_CCAAAAUACCCAUGACUCUU .....(((((((.(((((...((((................................................................))))..))))).)))))))....... (-14.42 = -14.78 + 0.36)


| Location | 10,765,323 – 10,765,428 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.71 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -15.34 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
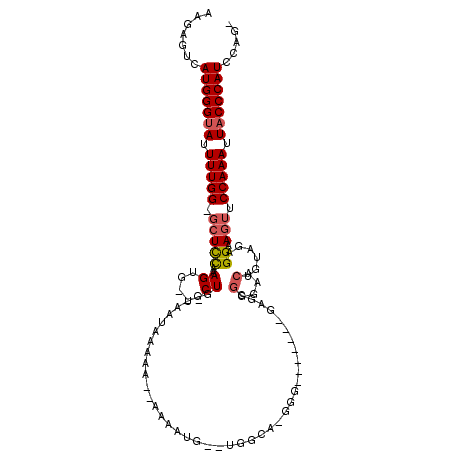
>2R_DroMel_CAF1 10765323 105 - 20766785 AAGAGUCAUGGGUAUUUUGG-GAUCCAAAGUG---CUGGUAAUACAAA--AAAAUG--UAGGG-GGGGGGGGAAGAGCGGGAACUGUAGAGGAAGUUCCAAAUUACCCAUCCAG- .......((.((((((((((-...))))))))---)).))..((((..--....))--))...-((..(((...((...((((((........))))))...)).)))..))..- ( -29.90) >DroSec_CAF1 1086 98 - 1 AAGAGUCAUGGGUAUUUUGG-GCUCCAAAGUG---CUGGUAAUAAAAA--AAAUUG--UGUCG-GGG-------GAGGGGGAACUGUAGAGGAAGUUCCAAAUUACCCAUCCAG- .......((.((((((((((-...))))))))---)).))........--....((--.(..(-((.-------.(...((((((........))))))...)..)))..))).- ( -27.20) >DroSim_CAF1 1164 101 - 1 AAGAGUCAUGGGUAUUUUGG-GCUCCAAAGUG---CUGGUAAUAAAAAAAAAAUUG--GGAGAGGGG-------AAGCGGGUACUGUAGAGGAAGUUCCAAAUUACCCAUCCAG- .......(((((((.(((((-(((((..((((---((.((................--.........-------..)).)))))).....)).)).)))))).)))))))....- ( -26.43) >DroEre_CAF1 1120 107 - 1 AAGAGUCAUGGGUAUUUUGG-CCUCCAAAGUG---CUGGUAAUAAAAA--AGAACGGACGGCA-GGGUUGGCAGGGGCGGGAACUGUAGAGGAAGUUCCAAAUUACCCAUCCAG- .......(((((((.(((.(-(((((((..((---(((..........--........)))))-...))))...)))).((((((........))))))))).)))))))....- ( -32.87) >DroYak_CAF1 1094 97 - 1 AAGAGUCAUGGGUAUUUUGG-GCUCCAAAGUG---CUGGUAAUAAAAA--AAA---GGUGGAA-GG-------GGAGCGGCAACUGUAGAGGAAGUUCCAAAUUACCCAUCCAG- .......(((((((.(((((-(((((.....(---((..(........--)..---)))....-..-------)))))...((((........))))))))).)))))))....- ( -26.10) >DroAna_CAF1 14867 89 - 1 AACAGUCAUGGGCAUUUUGGGGCUUUCAAGUACUACUGGCCAUUAAAA-----CUG---------------------GGGGAUACGUGGAGGAAGUACCAAAUUACCCAUCCAUC .......(((((...(((((..(((((..((((..(..(.........-----)..---------------------)..).))).....)))))..)))))...)))))..... ( -22.40) >consensus AAGAGUCAUGGGUAUUUUGG_GCUCCAAAGUG___CUGGUAAUAAAAA__AAAAUG__UGGCA_GGG_______GAGCGGGAACUGUAGAGGAAGUUCCAAAUUACCCAUCCAG_ .......(((((((.(((((.(((((..((.....)).........................................(....)......)).))).))))).)))))))..... (-15.34 = -15.73 + 0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:49 2006