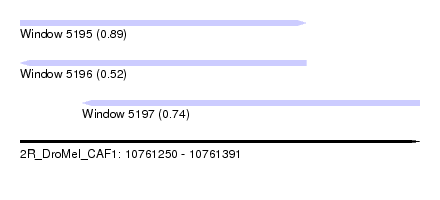
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,761,250 – 10,761,391 |
| Length | 141 |
| Max. P | 0.890043 |

| Location | 10,761,250 – 10,761,351 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -24.26 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.02 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10761250 101 + 20766785 ------GUAUAUAAAUUGGUGACGUCAUAGGAAGGUGAACACCUGCAUUGAUUCACGGACGUGUUUAGUUUUUU-UUUACACUUCCUUUUGACAUUUAUGCCUGCCCA ------(((((......(....)((((.(((((((((((.......(((((..(((....))).))))).....-))))).))))))..))))...)))))....... ( -23.50) >DroSec_CAF1 27184 102 + 1 ------GUUUAUAAAUUGGUGACGUCAUAGGAAGGUGAACACCUGCAUUGAUUCACGGACGUGUUUAGUUUUUUCUUUACACUUCCUUUUGACAUUUAUGCCUGCCCA ------...(((((((.(((((.((((...(.((((....)))).)..))))))))(((.((((..((......))..)))).)))......))))))))........ ( -23.70) >DroSim_CAF1 26805 101 + 1 ------GUAUAUAAAUUGGUGACGUCAUAGGAAGGUGAACACCUGCAUUGAUUCACGGACGUGUUUAGUUUUUU-UUUACACUUCCUUUUGACAUUUAUGCCUGCCCA ------(((((......(....)((((.(((((((((((.......(((((..(((....))).))))).....-))))).))))))..))))...)))))....... ( -23.50) >DroEre_CAF1 25266 99 + 1 ------GUAUGUACAUUGGUGACGUCAUAGGAAGGUGAACACCUGCAUUGAUUCACGGACGUGUUCAGUUU--U-UUUACACUUCCUUUUGACAUUUAUGCCUGCCCA ------(((.(((....(....)((((.((((((.((((((((((..........)))..)))))))((..--.-...)).))))))..)))).....))).)))... ( -25.10) >DroYak_CAF1 26090 106 + 1 AGGUACAUAUAUACAUUGGUGACGUCAUAGGAAGGUGAACACCUGCAUUGAUUCACGAACGUGUUUAGUUUU-U-UUUACACUUCCUUUUGACAUUUAUGCCUGCCCA (((((............(....)((((.(((((((((((.......(((((..(((....))).)))))...-.-))))).))))))..)))).....)))))..... ( -25.50) >consensus ______GUAUAUAAAUUGGUGACGUCAUAGGAAGGUGAACACCUGCAUUGAUUCACGGACGUGUUUAGUUUUUU_UUUACACUUCCUUUUGACAUUUAUGCCUGCCCA .................(((((.((((.(((((((((((.......(((((..(((....))).)))))......))))).))))))..)))).))...)))...... (-22.18 = -22.02 + -0.16)

| Location | 10,761,250 – 10,761,351 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -21.19 |
| Consensus MFE | -19.04 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10761250 101 - 20766785 UGGGCAGGCAUAAAUGUCAAAAGGAAGUGUAAA-AAAAAACUAAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCUAUGACGUCACCAAUUUAUAUAC------ .((((.((((....))))........((((...-..........))))))))((((.(((..(.((((....)))).)...)))..))))............------ ( -19.52) >DroSec_CAF1 27184 102 - 1 UGGGCAGGCAUAAAUGUCAAAAGGAAGUGUAAAGAAAAAACUAAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCUAUGACGUCACCAAUUUAUAAAC------ .((((.((((....))))........((((..((......))..))))))))((((.(((..(.((((....)))).)...)))..))))............------ ( -20.80) >DroSim_CAF1 26805 101 - 1 UGGGCAGGCAUAAAUGUCAAAAGGAAGUGUAAA-AAAAAACUAAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCUAUGACGUCACCAAUUUAUAUAC------ .((((.((((....))))........((((...-..........))))))))((((.(((..(.((((....)))).)...)))..))))............------ ( -19.52) >DroEre_CAF1 25266 99 - 1 UGGGCAGGCAUAAAUGUCAAAAGGAAGUGUAAA-A--AAACUGAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCUAUGACGUCACCAAUGUACAUAC------ ((..((.......((((((..((((((.((...-.--..))(((((((.(.(((......))).).))))))).)))))).))))))......))..))...------ ( -24.52) >DroYak_CAF1 26090 106 - 1 UGGGCAGGCAUAAAUGUCAAAAGGAAGUGUAAA-A-AAAACUAAACACGUUCGUGAAUCAAUGCAGGUGUUCACCUUCCUAUGACGUCACCAAUGUAUAUAUGUACCU .(((((.((((..((((((..((((((((....-.-.......(((((.(.(((......))).).))))))).)))))).)))))).....)))).....))).)). ( -21.60) >consensus UGGGCAGGCAUAAAUGUCAAAAGGAAGUGUAAA_AAAAAACUAAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCUAUGACGUCACCAAUUUAUAUAC______ .((((.((((....))))........((((..............))))))))((((.(((..(.((((....)))).)...)))..)))).................. (-19.04 = -18.88 + -0.16)

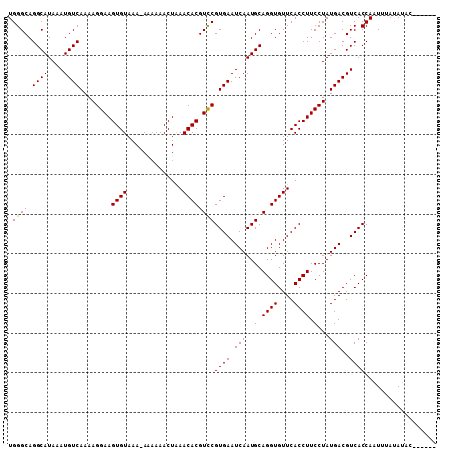
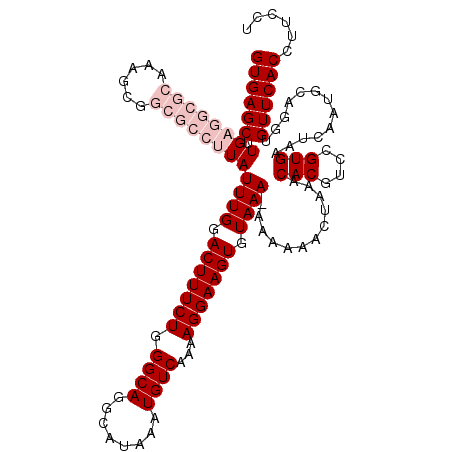
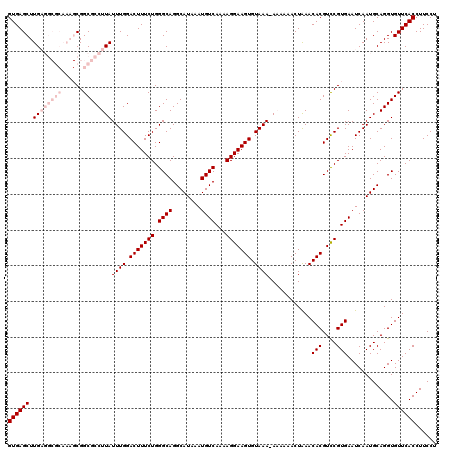
| Location | 10,761,272 – 10,761,391 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.07 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -26.64 |
| Energy contribution | -29.04 |
| Covariance contribution | 2.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10761272 119 - 20766785 GUGAGCUUGAGGCGCAAAGCGGCGCCUUAUUUGGACUUUCUGGGCAGGCAUAAAUGUCAAAAGGAAGUGUAAA-AAAAAACUAAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCU ((((((.((((((((......))))))))((((.(((((((.((((........))))...))))))).))))-...........(((....))).............))))))...... ( -36.60) >DroSec_CAF1 27206 120 - 1 GUGAGCUUGAGGCGCAAAGCGGCGCCUUAUUUGGACUUUCUGGGCAGGCAUAAAUGUCAAAAGGAAGUGUAAAGAAAAAACUAAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCU ((((((.((((((((......)))))))).(..(.....)..)((..((((.....(((...(((.((((..((......))..)))).))).)))....))))..))))))))...... ( -37.10) >DroSim_CAF1 26827 119 - 1 GUGAGCUUGAGGCGCAAAGCGGCGCCUUAUUUGGACUUUCUGGGCAGGCAUAAAUGUCAAAAGGAAGUGUAAA-AAAAAACUAAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCU ((((((.((((((((......))))))))((((.(((((((.((((........))))...))))))).))))-...........(((....))).............))))))...... ( -36.60) >DroEre_CAF1 25288 107 - 1 GUGAGCUUG----------CGGCGCCUUAUUUGGACUUUCUGGGCAGGCAUAAAUGUCAAAAGGAAGUGUAAA-A--AAACUGAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCU (((((((((----------((........((((.(((((((.((((........))))...))))))).))))-.--....(((.(((....)))..))).)))))))..))))...... ( -28.50) >DroYak_CAF1 26118 108 - 1 GUGAGCUUG----------CGGCGCCUUAUUUGGACUUUCUGGGCAGGCAUAAAUGUCAAAAGGAAGUGUAAA-A-AAAACUAAACACGUUCGUGAAUCAAUGCAGGUGUUCACCUUCCU (((((((((----------((........((((.(((((((.((((........))))...))))))).))))-.-.........(((....)))......)))))))..))))...... ( -28.40) >consensus GUGAGCUUGAGGCGCAAAGCGGCGCCUUAUUUGGACUUUCUGGGCAGGCAUAAAUGUCAAAAGGAAGUGUAAA_AAAAAACUAAACACGUCCGUGAAUCAAUGCAGGUGUUCACCUUCCU ((((((.((((((((......))))))))((((.(((((((.((((........))))...))))))).))))............(((....))).............))))))...... (-26.64 = -29.04 + 2.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:47 2006