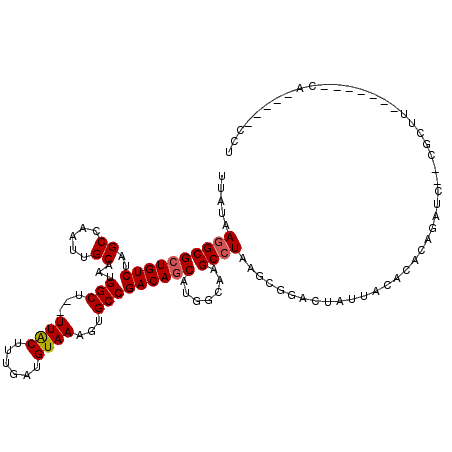
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,759,186 – 10,759,301 |
| Length | 115 |
| Max. P | 0.796503 |

| Location | 10,759,186 – 10,759,301 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -19.79 |
| Energy contribution | -20.02 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10759186 115 + 20766785 AGGGGCUGCGGUGGAGCAAGCG--GCUCUGUGUGUAAUAGUCCGCUUAGGCUUGCCAUGCUGUCGGCACUUUACAUCAAAGUAA--AGCCAUUGCAAUUGGCUAGACAGCGCCUUAUAA .(((((...((..(.((.((((--(..((((.....)))).)))))...)))..))..((((((...(((((.....)))))..--(((((.......))))).))))))))))).... ( -44.60) >DroEre_CAF1 23198 115 + 1 AGGGGCUGUGGAGGGGCAAGCG--GAUCUGUGUGUAAUAGUCCGCUUAGGCUUGCCAUGCUGUCGGCACUUUACAUCAAAGUAA--AGCCAUUGCAAUUGGCUAGACAGCGCCUUAUAA .(((((.((((..((.((((((--((.((((.....))))))))))).).))..))))((((((...(((((.....)))))..--(((((.......))))).))))))))))).... ( -49.00) >DroWil_CAF1 30755 113 + 1 AGGGGUGGUG------UGCGUGGUCAUGGGUGAGUAAUAGUCCGCUUAGGCUUGUUCUGCUGUCGGCACUUUACAUCAAAGCAAAAAGCCAUUGCAAUUGGCUAGACAUCGCCUUAUAA .(((((((((------(.(.((((((....(((((((.((((((....(((.......)))..))).))))))).)))..((((.......))))...))))))))))))))))).... ( -34.40) >DroYak_CAF1 23873 109 + 1 UGG-----UGCAA-GGGAAGCG--GAUCUGUGUGUAAUAGUCCGCUUAGGCUUGCCAUGCUGUCGGCACUUUACAUCAAAGUAA--AGCCAUUGCAAUUGGCUAGACAGCGCCUUAUAA .((-----(((((-(..(((((--((.((((.....)))))))))))...)))))...((((((...(((((.....)))))..--(((((.......))))).)))))))))...... ( -42.60) >DroAna_CAF1 21404 99 + 1 UUU-----UG-----------U--CUUCUGUGUGUAAUAGUCCGCUUAGGCUUGCCAUGCUGUCGGCACUUUACAUCAAAGUAA--AGCCAUUGCAAUUGGCUAGACAGCGCCUUAUAA ..(-----((-----------(--((..((.((((((.((((((....(((.......)))..))).)))))))))))......--(((((.......))))))))))).......... ( -24.20) >DroPer_CAF1 22199 97 + 1 -------------------CCG--GCCGUGUGUGGCACAGUCCGCUUAGGCUUUCUUGGCUGUCGGCACUUUACAUCAAAGUAA-AAGCCAUUGCGAUUGGCUAGACAGCGCUUUAUAA -------------------..(--(((....((((......))))...))))......((((((.....(((((......))))-)(((((.......))))).))))))......... ( -29.70) >consensus AGG_____UG_______AAGCG__GAUCUGUGUGUAAUAGUCCGCUUAGGCUUGCCAUGCUGUCGGCACUUUACAUCAAAGUAA__AGCCAUUGCAAUUGGCUAGACAGCGCCUUAUAA .............................((((((((.((((((....(((.......)))..))).)))))))......((....(((((.......)))))..)).))))....... (-19.79 = -20.02 + 0.22)

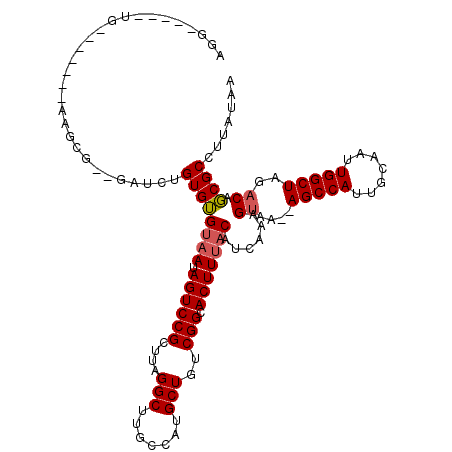
| Location | 10,759,186 – 10,759,301 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.49 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10759186 115 - 20766785 UUAUAAGGCGCUGUCUAGCCAAUUGCAAUGGCU--UUACUUUGAUGUAAAGUGCCGACAGCAUGGCAAGCCUAAGCGGACUAUUACACACAGAGC--CGCUUGCUCCACCGCAGCCCCU ......(((((((((..((.....))....(((--((((......)))))))...)))))).(((..(((..((((((.((...........)))--))))))))))).....)))... ( -36.10) >DroEre_CAF1 23198 115 - 1 UUAUAAGGCGCUGUCUAGCCAAUUGCAAUGGCU--UUACUUUGAUGUAAAGUGCCGACAGCAUGGCAAGCCUAAGCGGACUAUUACACACAGAUC--CGCUUGCCCCUCCACAGCCCCU ......(((((((((..((.....))....(((--((((......)))))))...))))))..((((......((((((((.........)).))--))))))))........)))... ( -35.90) >DroWil_CAF1 30755 113 - 1 UUAUAAGGCGAUGUCUAGCCAAUUGCAAUGGCUUUUUGCUUUGAUGUAAAGUGCCGACAGCAGAACAAGCCUAAGCGGACUAUUACUCACCCAUGACCACGCA------CACCACCCCU .....((((....(((.((.....))..((((.((((((......)))))).)))).....)))....))))..(((......................))).------.......... ( -22.25) >DroYak_CAF1 23873 109 - 1 UUAUAAGGCGCUGUCUAGCCAAUUGCAAUGGCU--UUACUUUGAUGUAAAGUGCCGACAGCAUGGCAAGCCUAAGCGGACUAUUACACACAGAUC--CGCUUCCC-UUGCA-----CCA ......((.((((((..((.....))....(((--((((......)))))))...))))))...(((((...(((((((((.........)).))--)))))..)-)))).-----)). ( -36.30) >DroAna_CAF1 21404 99 - 1 UUAUAAGGCGCUGUCUAGCCAAUUGCAAUGGCU--UUACUUUGAUGUAAAGUGCCGACAGCAUGGCAAGCCUAAGCGGACUAUUACACACAGAAG--A-----------CA-----AAA ...........((((((((((.......)))))--...((.((.((((((((.(((.......((....))....)))))).))))))).)).))--)-----------))-----... ( -22.70) >DroPer_CAF1 22199 97 - 1 UUAUAAAGCGCUGUCUAGCCAAUCGCAAUGGCUU-UUACUUUGAUGUAAAGUGCCGACAGCCAAGAAAGCCUAAGCGGACUGUGCCACACACGGC--CGG------------------- .......(.((((((..((.....))...(((((-((((......)))))).)))))))))).............(((.(((((.....))))))--)).------------------- ( -30.40) >consensus UUAUAAGGCGCUGUCUAGCCAAUUGCAAUGGCU__UUACUUUGAUGUAAAGUGCCGACAGCAUGGCAAGCCUAAGCGGACUAUUACACACAGAUC__CGCUU_______CA_____CCU .....((((((((((..((.....))...(((...((((......))))...))))))))).......))))............................................... (-20.30 = -20.49 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:45 2006