
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
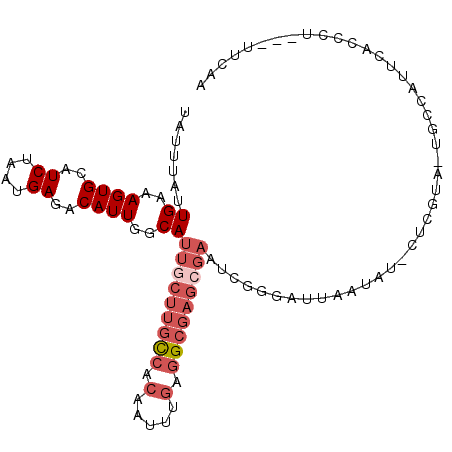
| Location | 10,758,293 – 10,758,422 |
| Length | 129 |
| Max. P | 0.615937 |

| Location | 10,758,293 – 10,758,394 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -12.44 |
| Energy contribution | -13.97 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
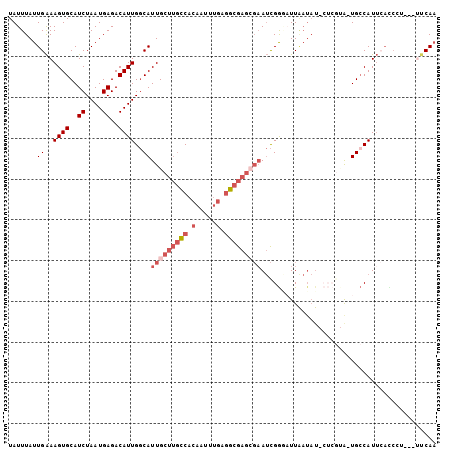
>2R_DroMel_CAF1 10758293 101 + 20766785 UAUUUAUUGAAAGUGCAUCUAAUGAGACAUUGGCAUUGCUUGCCACAAUUUGAGGCGAGCGAAUCGGGAUUAAUAUACUCGUA-UGCCAUUCACCCU---UUCAA ......(((((((((..((....))..)).((((((((((((((.(.....).)))))))))..((((((....)).))))..-)))))......))---))))) ( -28.60) >DroSec_CAF1 24196 101 + 1 UAUUUAUUGAAAGUGCAUCUAAUGAGACAUUGGCAUUGCUUGCCACAAUUUGAGGCGAGAGAAUCGGAAUUAAUAUACUCGUA-UGCCAUUCACCCU---UUCAA .......((((.(.((((...(((((....(((((.....)))))...(((((..(....)..))))).........))))))-)))).))))....---..... ( -22.60) >DroSim_CAF1 23803 93 + 1 UAUUUAUUGAAAGUGCAUCUAAUGAGACAUUGGCAUUGCUUGCCACAAUUUGAGGCGAGCGAAUCGGCAUUAAUAUACUCGUA-UGCCAUU-----------CAA .......((..((((..((....))..))))..))..(((((((.(.....).)))))))((((.(((((............)-)))))))-----------).. ( -26.10) >DroEre_CAF1 22244 95 + 1 UAUUUAUUGAAAGUGCAUCUAAUGAGACAUUGGCAUUGCUUGCCACAAUUUGAGGCGAGCGCAUCGGGCUUGAUAU------G-UGCCAUUCACCCU---UUCAA ......((((((((((..((((((...))))))....(((((((.(.....).))))))))))...(((.......------.-.))).......))---))))) ( -27.00) >DroYak_CAF1 22951 95 + 1 UAUUUAUUGAAAGUGCAUCUAAUGAGACAUUGGCAUUGCUUGCCACAAUUUGAGGCGAGCGAAUCGAGCUUGAUAU------A-UGACAUUCAUCCA---CUCAA .......((((.((.(((.((.((((...((((..(((((((((.(.....).))))))))).)))).)))).)).------)-)))).))))....---..... ( -22.40) >DroAna_CAF1 20779 94 + 1 UAUUUAUUGGAAGUGCAUCUAAUGAGACAUUGCCAUUGCUGCUCAUAAUU----GCGCCA-AAAUGGCAUU------CUCGCGCUGCCAUGAAUCAUUGAUUCAU .......(((.(((((.......((((...(((((((...((.((....)----).))..-.))))))).)------)))))))).)))((((((...)))))). ( -28.81) >consensus UAUUUAUUGAAAGUGCAUCUAAUGAGACAUUGGCAUUGCUUGCCACAAUUUGAGGCGAGCGAAUCGGGAUUAAUAU_CUCGUA_UGCCAUUCACCCU___UUCAA .......((..((((..((....))..))))..))(((((((((.(.....).)))))))))........................................... (-12.44 = -13.97 + 1.53)

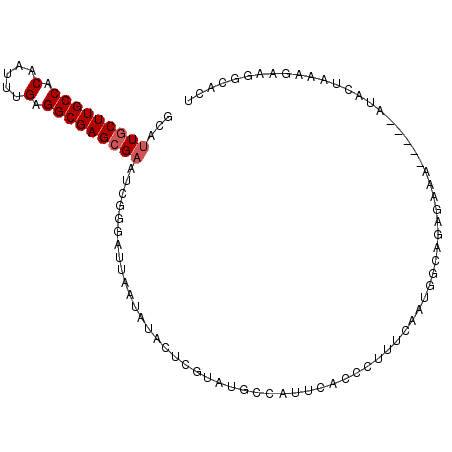
| Location | 10,758,325 – 10,758,422 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 73.88 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -12.16 |
| Energy contribution | -12.56 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
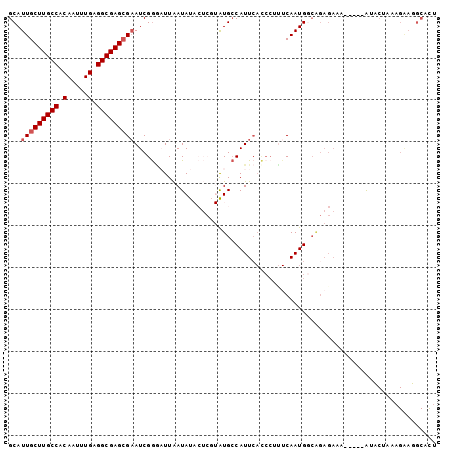
>2R_DroMel_CAF1 10758325 97 + 20766785 GCAUUGCUUGCCACAAUUUGAGGCGAGCGAAUCGGGAUUAAUAUACUCGUAUGCCAUUCACCCUUUCAAUGGCAGAGAAA-----AUUCUAAAGGAGGCAUU ((.(((((((((.(.....).)))))))))...............(((...(((((((.........))))))).(((..-----..)))....)))))... ( -26.60) >DroSec_CAF1 24228 83 + 1 GCAUUGCUUGCCACAAUUUGAGGCGAGAGAAUCGGAAUUAAUAUACUCGUAUGCCAUUCACCCUUUCAAUGCCAGAGAAA-----AU--------------U ((((((((((((.(.....).)))))).((((.((.....((((....)))).)))))).......))))))........-----..--------------. ( -18.80) >DroSim_CAF1 23835 89 + 1 GCAUUGCUUGCCACAAUUUGAGGCGAGCGAAUCGGCAUUAAUAUACUCGUAUGCCAUU--------CAAUGCCAGAGAAA-----AUUCUAAAGGAGGCAUU .....(((((((.(.....).)))))))((((.(((((............))))))))--------)((((((..(((..-----..)))......)))))) ( -27.00) >DroEre_CAF1 22276 83 + 1 GCAUUGCUUGCCACAAUUUGAGGCGAGCGCAUCGGGCUUGAUAU------GUGCCAUUCACCCUUUCAAUGG-------------GUACUAAAGAAUGCCCU ((((((((((((.(.....).)))))))......(((.......------..)))....((((.......))-------------)).......)))))... ( -25.70) >DroYak_CAF1 22983 96 + 1 GCAUUGCUUGCCACAAUUUGAGGCGAGCGAAUCGAGCUUGAUAU------AUGACAUUCAUCCACUCAAUGGCGGAGAAAAAAGAAUACUAAAGAAUGCCCU ((((((((((((.(.....).)))))))................------......(((.(((.((....)).))))))...............)))))... ( -21.50) >consensus GCAUUGCUUGCCACAAUUUGAGGCGAGCGAAUCGGGAUUAAUAUACUCGUAUGCCAUUCACCCUUUCAAUGGCAGAGAAA_____AUACUAAAGAAGGCACU ...(((((((((.(.....).)))))))))........................................................................ (-12.16 = -12.56 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:42 2006