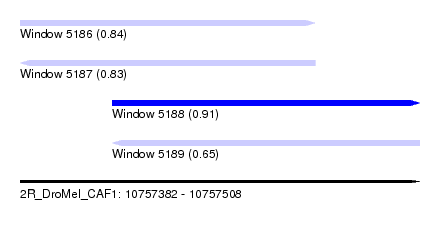
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,757,382 – 10,757,508 |
| Length | 126 |
| Max. P | 0.913966 |

| Location | 10,757,382 – 10,757,475 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -31.29 |
| Consensus MFE | -18.07 |
| Energy contribution | -18.15 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10757382 93 + 20766785 --------GUUAAAGUUCUUC---AGUGCAGCAUAAUGUGUUUUAAUCGCUGCGACAUGCUGCGC---UUUUGUGCUUACGACCGCAACAUGUUGCAAGUUGGAAUU------------- --------.....((((((..---..((((((((...(((.......)))((((.......((((---....)))).......))))..))))))))....))))))------------- ( -25.64) >DroSec_CAF1 23287 103 + 1 --------AUUAAAGUUCUUC---AG--UAGCAUAAGGGUUUUUAAUCGCUGCGACAUGUUGCGC---UUUUGUGCUUACGACCGCAACAUGUUGCA-GUUGGAAUUGUUUAAACCGAAG --------.........((((---..--...(....)(((((.(((((((((((((((((((((.---..((((....)))).))))))))))))))-))..).))))...))))))))) ( -32.30) >DroSim_CAF1 22851 95 + 1 --------AUUAAAGUUCUUC---AG--CAGCAUAAGGGUUUUUAAUCGCUGCGACAUGUUGCGC-----------UUACGACCGCAACAUGUUGCA-GUUGGAAUUGUUUAAACCGAAG --------.........((((---..--...(....)(((((.(((((((((((((((((((((.-----------.......))))))))))))))-))..).))))...))))))))) ( -31.30) >DroEre_CAF1 21181 114 + 1 GCAUU-UCAUAAAAGUUCUUC---AGUGCUGCAUGAGGGCUUUUCAUCGCUGCGACGUGCUGCGCAACUUUUGUGCUUGCGACCGCAACAUGUUGCA-GUUGGAUUUGUUUAAAGCG-AG (((..-((..((((((.((((---(........)))))))))))....(((((((((((..(((((.....)))))((((....)))))))))))))-))..))..)))........-.. ( -34.90) >DroYak_CAF1 21929 119 + 1 GGAUUUUUAUAAAAGUUCUGCAUGAGUGCUGCAUAAGGGGUUUUAAUCGCUGCGACAUGCUGUGCAACUUUUGUGCUUGCGACCGCAACAUGUUGCA-GUUGGAAUUGUUUAAAACGAAG (((((((....)))))))((((.......))))((((.(((((((...(((((((((((..(..((.....))..)((((....)))))))))))))-))))))))).))))........ ( -32.30) >consensus ________AUUAAAGUUCUUC___AGUGCAGCAUAAGGGGUUUUAAUCGCUGCGACAUGCUGCGC___UUUUGUGCUUACGACCGCAACAUGUUGCA_GUUGGAAUUGUUUAAACCGAAG ........................................(((((((...(((((((((.((((...................)))).))))))))).)))))))............... (-18.07 = -18.15 + 0.08)

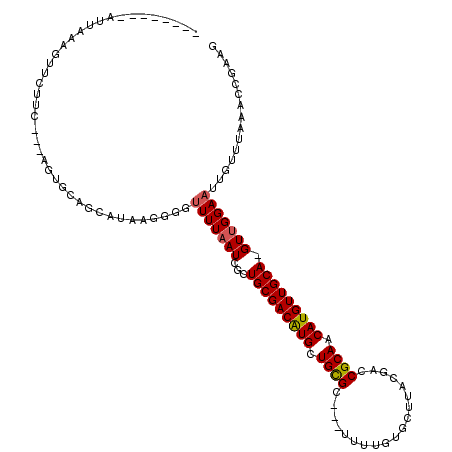
| Location | 10,757,382 – 10,757,475 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -15.49 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10757382 93 - 20766785 -------------AAUUCCAACUUGCAACAUGUUGCGGUCGUAAGCACAAAA---GCGCAGCAUGUCGCAGCGAUUAAAACACAUUAUGCUGCACU---GAAGAACUUUAAC-------- -------------........(((.(((((((((((((((....).))....---.)))))))))).((((((..(((......)))))))))..)---)))).........-------- ( -22.60) >DroSec_CAF1 23287 103 - 1 CUUCGGUUUAAACAAUUCCAAC-UGCAACAUGUUGCGGUCGUAAGCACAAAA---GCGCAACAUGUCGCAGCGAUUAAAAACCCUUAUGCUA--CU---GAAGAACUUUAAU-------- (((((((..............(-(((.(((((((((((((....).))....---.)))))))))).))))....(((......)))....)--))---)))).........-------- ( -26.30) >DroSim_CAF1 22851 95 - 1 CUUCGGUUUAAACAAUUCCAAC-UGCAACAUGUUGCGGUCGUAA-----------GCGCAACAUGUCGCAGCGAUUAAAAACCCUUAUGCUG--CU---GAAGAACUUUAAU-------- (((((((..............(-(((.((((((((((.......-----------.)))))))))).))))....(((......)))....)--))---)))).........-------- ( -27.00) >DroEre_CAF1 21181 114 - 1 CU-CGCUUUAAACAAAUCCAAC-UGCAACAUGUUGCGGUCGCAAGCACAAAAGUUGCGCAGCACGUCGCAGCGAUGAAAAGCCCUCAUGCAGCACU---GAAGAACUUUUAUGA-AAUGC ..-.(((((......(((...(-(((.((.((((((((.(....))((....))..))))))).)).)))).)))..)))))......(((.((..---(((....)))..)).-..))) ( -23.60) >DroYak_CAF1 21929 119 - 1 CUUCGUUUUAAACAAUUCCAAC-UGCAACAUGUUGCGGUCGCAAGCACAAAAGUUGCACAGCAUGUCGCAGCGAUUAAAACCCCUUAUGCAGCACUCAUGCAGAACUUUUAUAAAAAUCC .....................(-((((...((((((((((....).))....(((((((.....)).)))))...............)))))))....)))))................. ( -24.60) >consensus CUUCGGUUUAAACAAUUCCAAC_UGCAACAUGUUGCGGUCGUAAGCACAAAA___GCGCAGCAUGUCGCAGCGAUUAAAAACCCUUAUGCUGCACU___GAAGAACUUUAAU________ .......................(((.((((((((((...................)))))))))).))).................................................. (-15.49 = -15.65 + 0.16)


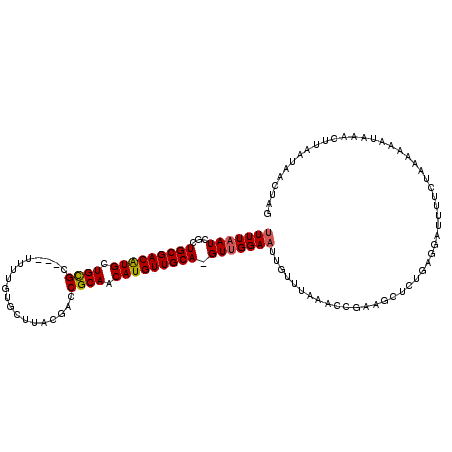
| Location | 10,757,411 – 10,757,508 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -17.91 |
| Energy contribution | -17.99 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10757411 97 + 20766785 UUUUAAUCGCUGCGACAUGCUGCGC---UUUUGUGCUUACGACCGCAACAUGUUGCAAGUUGGAAUU-----------------GAAGAUUUCCUAAAAAAUAAACUUAAUAACUAG (((((...(((((((((((.((((.---..((((....)))).)))).)))))))).))).((((..-----------------......))))))))).................. ( -22.70) >DroSec_CAF1 23314 113 + 1 UUUUAAUCGCUGCGACAUGUUGCGC---UUUUGUGCUUACGACCGCAACAUGUUGCA-GUUGGAAUUGUUUAAACCGAAGCUCUGAGGAUUUUCGAAAAAAUAAACUUAAUAACUAG ........((((((((((((((((.---..((((....)))).))))))))))))))-)).....((((((....(((((.((....)).)))))...))))))............. ( -32.80) >DroSim_CAF1 22878 105 + 1 UUUUAAUCGCUGCGACAUGUUGCGC-----------UUACGACCGCAACAUGUUGCA-GUUGGAAUUGUUUAAACCGAAGCUCUGAGGAUUUUCGACAAAAUAAACUUAAUAACUAG ........((((((((((((((((.-----------.......))))))))))))))-)).....((((((....(((((.((....)).)))))...))))))............. ( -31.80) >DroEre_CAF1 21217 108 + 1 UUUUCAUCGCUGCGACGUGCUGCGCAACUUUUGUGCUUGCGACCGCAACAUGUUGCA-GUUGGAUUUGUUUAAAGCG-AGCUCUGAGGAAUUUCUAAAAAAUAAUCUUGC------- ((((((..(((((((((((..(((((.....)))))((((....)))))))))))))-)).(..(((((.....)))-))..))))))).....................------- ( -30.00) >DroYak_CAF1 21969 116 + 1 UUUUAAUCGCUGCGACAUGCUGUGCAACUUUUGUGCUUGCGACCGCAACAUGUUGCA-GUUGGAAUUGUUUAAAACGAAGCUCUGGGGAUUUUUUAAAAAAUAUUCUUGUUAAAUAG .((((((.(((((((((((..(..((.....))..)((((....)))))))))))))-)).(((((..(((((((.....((....))...)))))))....))))).))))))... ( -28.00) >consensus UUUUAAUCGCUGCGACAUGCUGCGC___UUUUGUGCUUACGACCGCAACAUGUUGCA_GUUGGAAUUGUUUAAACCGAAGCUCUGAGGAUUUUCUAAAAAAUAAACUUAAUAACUAG (((((((...(((((((((.((((...................)))).))))))))).))))))).................................................... (-17.91 = -17.99 + 0.08)

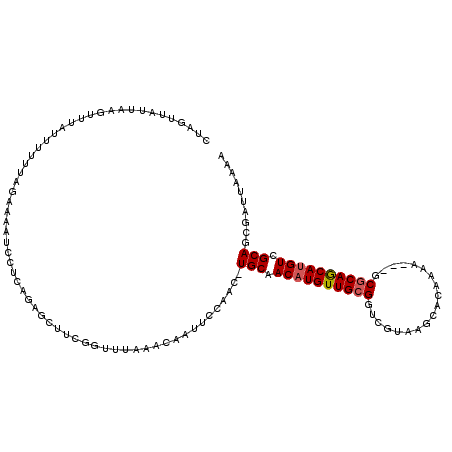
| Location | 10,757,411 – 10,757,508 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -24.81 |
| Consensus MFE | -15.49 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10757411 97 - 20766785 CUAGUUAUUAAGUUUAUUUUUUAGGAAAUCUUC-----------------AAUUCCAACUUGCAACAUGUUGCGGUCGUAAGCACAAAA---GCGCAGCAUGUCGCAGCGAUUAAAA .(((((.................((((......-----------------..))))...((((.(((((((((((((....).))....---.)))))))))).)))).)))))... ( -23.10) >DroSec_CAF1 23314 113 - 1 CUAGUUAUUAAGUUUAUUUUUUCGAAAAUCCUCAGAGCUUCGGUUUAAACAAUUCCAAC-UGCAACAUGUUGCGGUCGUAAGCACAAAA---GCGCAACAUGUCGCAGCGAUUAAAA .(((((.....(((((.....(((((..((....))..)))))..)))))........(-(((.(((((((((((((....).))....---.)))))))))).)))).)))))... ( -29.30) >DroSim_CAF1 22878 105 - 1 CUAGUUAUUAAGUUUAUUUUGUCGAAAAUCCUCAGAGCUUCGGUUUAAACAAUUCCAAC-UGCAACAUGUUGCGGUCGUAA-----------GCGCAACAUGUCGCAGCGAUUAAAA ................(((.((((..........((((....))))............(-(((.((((((((((.......-----------.)))))))))).)))))))).))). ( -29.40) >DroEre_CAF1 21217 108 - 1 -------GCAAGAUUAUUUUUUAGAAAUUCCUCAGAGCU-CGCUUUAAACAAAUCCAAC-UGCAACAUGUUGCGGUCGCAAGCACAAAAGUUGCGCAGCACGUCGCAGCGAUGAAAA -------....................(((.((...(((-.(((((....)))....((-(((((....))))))).)).)))......(((((((.....).)))))))).))).. ( -22.20) >DroYak_CAF1 21969 116 - 1 CUAUUUAACAAGAAUAUUUUUUAAAAAAUCCCCAGAGCUUCGUUUUAAACAAUUCCAAC-UGCAACAUGUUGCGGUCGCAAGCACAAAAGUUGCACAGCAUGUCGCAGCGAUUAAAA ..........................((((......((((((...............((-(((((....))))))))).))))......(((((((.....)).))))))))).... ( -20.06) >consensus CUAGUUAUUAAGUUUAUUUUUUAGAAAAUCCUCAGAGCUUCGGUUUAAACAAUUCCAAC_UGCAACAUGUUGCGGUCGUAAGCACAAAA___GCGCAGCAUGUCGCAGCGAUUAAAA ............................................................(((.((((((((((...................)))))))))).))).......... (-15.49 = -15.65 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:40 2006