| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,750,472 – 10,750,570 |
| Length | 98 |
| Max. P | 0.668301 |

| Location | 10,750,472 – 10,750,570 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.70 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -20.98 |
| Energy contribution | -21.93 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
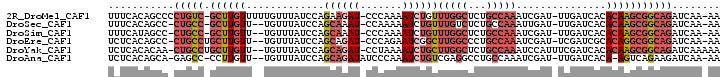
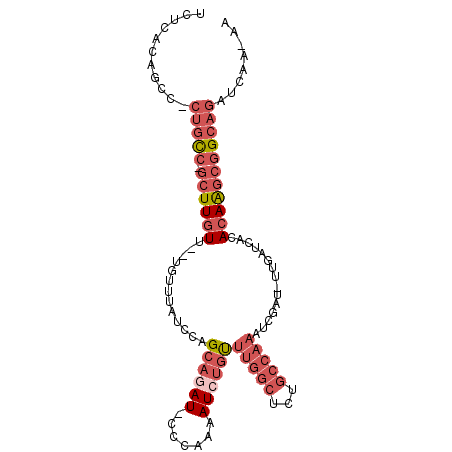
>2R_DroMel_CAF1 10750472 98 + 20766785 UU-UUGAUCUGCCGCUUGUGUGAUCAA-AUCGAUUUGGCAGAGCCAAACAGAUUUUGGG-AUCUUCUGGAUAAACAAAAACAAGC-GACAGGGGCUGUGAAA ..-....((((.(((((((..((((((-(((..((((((...))))))..)))))...)-)))...((......))...))))))-).)))).......... ( -27.20) >DroSec_CAF1 14800 95 + 1 UU-UUGAUCUGCCGCUUGUGUGAUCAA-AUCAAUUUGGCAGAGACAAACAGAUUUUUGG-AUUUGCUGGAUAAACA--AACAAGC-GGCAG-GGCUGUGAAA ..-....((((((((((((.((((...-))))(((..(((((..((((......)))).-.)))))..))).....--.))))))-)))))-)......... ( -35.50) >DroSim_CAF1 15969 95 + 1 UU-UUGAUCUGCCGCUUGUGUGAUCAA-AUCGAUUUGGCAGAGCCAAACAGAUUUUGGG-AUUUGCUGGAUAAACA--AACAAGC-GGCAG-GGCUAUGAAA ..-....((((((((((((.((((...-))))(((..(((((.(((((.....))))).-.)))))..))).....--.))))))-)))))-)......... ( -35.30) >DroEre_CAF1 14412 96 + 1 UU-UUGAUCUGCCGCCUGUGCGAUCGA-AUCGAUUUGGCAGGGCCAAGCCGAUUCUGGG-AUCUGCUGGAUAAACA--AACAAGCAGGCAG-GGCUGUGAGA ..-....((.(((((((((((((((((-((((..(((((...)))))..))))))...)-))).))((......))--.....))))))..-)))...)).. ( -33.10) >DroYak_CAF1 14705 98 + 1 UUUUUGAUCUGCCGCUUGUGUGAUCGAAAUGGAUUUGGCAGAGCCAAGCAGAUUUUAGG-AUCUGCUGGAUAAACA--AACAAGCAGGCAG-UUGUGUGAGA ........(((((((((((...(((..........((((...))))((((((((....)-))))))).))).....--.)))))).)))))-.......... ( -32.00) >DroAna_CAF1 12836 95 + 1 UU-UUGAUCUUCUGACU-UGUGAUCAA-AUCGAUUUGGCAGGCCUCGACAGAUUUGGGAUAUCUGCUGGAUAAACA--AACAAGG-GGCUC-UGCUGUGAGA .(-((((((........-...))))))-)(((....(((((((((((.(((((.......))))))((......))--.....))-))).)-)))).))).. ( -26.30) >consensus UU_UUGAUCUGCCGCUUGUGUGAUCAA_AUCGAUUUGGCAGAGCCAAACAGAUUUUGGG_AUCUGCUGGAUAAACA__AACAAGC_GGCAG_GGCUGUGAAA ........(((((((((((.((((....))))(((..(((((.(((((.....)))))...)))))..)))........)))))).)))))........... (-20.98 = -21.93 + 0.95)

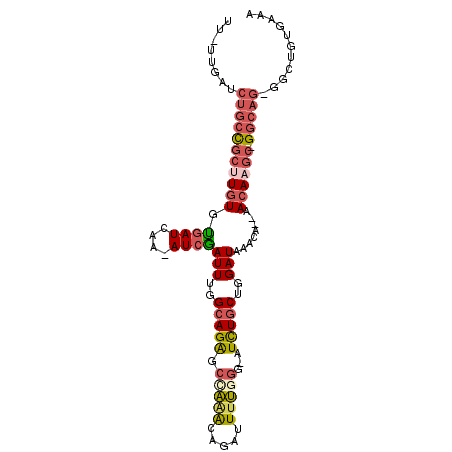
| Location | 10,750,472 – 10,750,570 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.70 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -13.50 |
| Energy contribution | -15.58 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.23 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10750472 98 - 20766785 UUUCACAGCCCCUGUC-GCUUGUUUUUGUUUAUCCAGAAGAU-CCCAAAAUCUGUUUGGCUCUGCCAAAUCGAU-UUGAUCACACAAGCGGCAGAUCAA-AA ...........(((((-(((((((((((......)))))(((-(...(((((.(((((((...))))))).)))-))))))..))))))))))).....-.. ( -33.60) >DroSec_CAF1 14800 95 - 1 UUUCACAGCC-CUGCC-GCUUGUU--UGUUUAUCCAGCAAAU-CCAAAAAUCUGUUUGUCUCUGCCAAAUUGAU-UUGAUCACACAAGCGGCAGAUCAA-AA ..........-(((((-(((((((--((((.....)))))..-....(((((.(((((.......))))).)))-))......))))))))))).....-.. ( -26.90) >DroSim_CAF1 15969 95 - 1 UUUCAUAGCC-CUGCC-GCUUGUU--UGUUUAUCCAGCAAAU-CCCAAAAUCUGUUUGGCUCUGCCAAAUCGAU-UUGAUCACACAAGCGGCAGAUCAA-AA ..........-(((((-(((((((--((((.....)))))..-....(((((.(((((((...))))))).)))-))......))))))))))).....-.. ( -32.80) >DroEre_CAF1 14412 96 - 1 UCUCACAGCC-CUGCCUGCUUGUU--UGUUUAUCCAGCAGAU-CCCAGAAUCGGCUUGGCCCUGCCAAAUCGAU-UCGAUCGCACAGGCGGCAGAUCAA-AA ..........-(((((.((((((.--((......))((.(((-(...((((((..(((((...)))))..))))-))))))))))))))))))).....-.. ( -35.40) >DroYak_CAF1 14705 98 - 1 UCUCACACAA-CUGCCUGCUUGUU--UGUUUAUCCAGCAGAU-CCUAAAAUCUGCUUGGCUCUGCCAAAUCCAUUUCGAUCACACAAGCGGCAGAUCAAAAA ..........-(((((.((((((.--((...(((..((((((-......))))))(((((...))))).........))))).)))))))))))........ ( -30.20) >DroAna_CAF1 12836 95 - 1 UCUCACAGCA-GAGCC-CCUUGUU--UGUUUAUCCAGCAGAUAUCCCAAAUCUGUCGAGGCCUGCCAAAUCGAU-UUGAUCACA-AGUCAGAAGAUCAA-AA ((((...(((-(.(((-.(.....--((......))((((((.......)))))).).)))))))......(((-(((....))-)))).).)))....-.. ( -19.70) >consensus UCUCACAGCC_CUGCC_GCUUGUU__UGUUUAUCCAGCAGAU_CCCAAAAUCUGUUUGGCUCUGCCAAAUCGAU_UUGAUCACACAAGCGGCAGAUCAA_AA ...........(((((.((((((.............((((((.......))))))(((((...)))))...............)))))))))))........ (-13.50 = -15.58 + 2.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:33 2006