
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
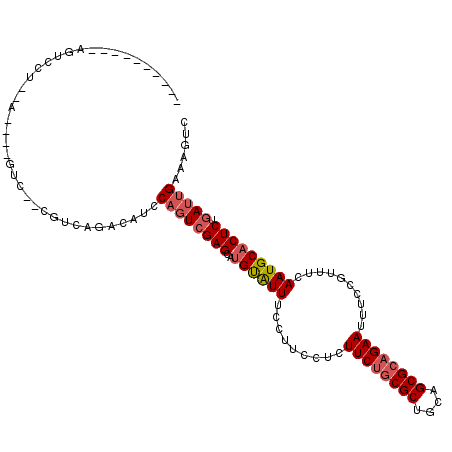
| Location | 10,684,823 – 10,684,937 |
| Length | 114 |
| Max. P | 0.924942 |

| Location | 10,684,823 – 10,684,917 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.35 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -19.67 |
| Energy contribution | -20.39 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10684823 94 + 20766785 -------------CCA--C----GUC--CGUCAGACAUCCAGUCGAGCAUGCAUUUCCUUCCUCUUCUGCGCUGCAGCGCAGAAUUUCCAUUCCAAUGCACUCUGAUUGAAAGUC -------------...--.----(((--.....)))...((((((((..((((((.........((((((((....))))))))..........))))))))).)))))...... ( -27.01) >DroEre_CAF1 22005 94 + 1 -------------CCA--C----GUC--CGUCAGACAUCCAGUCGAGCAUGUAUUUCCUUCCUCUUCCACGCUGCAGCGCAGAAUUUCCAUUCCAAUGCACUCUGAUUGAAAGUC -------------...--.----(((--.....)))...((((((((..((((((.........(((..(((....)))..)))..........))))))))).)))))...... ( -16.41) >DroWil_CAF1 31302 109 + 1 UUACUCACUCAGUCACUGA----GGC--AUUCAGUCAGCCAGUCGAGCAUGUGUUUCCUUCCUUUUCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCCCUCUGAUUGAAAGAC ....((..(((((((..(.----(((--(((.((..(((((........)).)))..)).....((((((((....))))))))..........)))))))..)))))))..)). ( -30.70) >DroMoj_CAF1 37437 104 + 1 --------UCAGUCAUUCACUCAUUC--UGUCAGCCAUUCCCUCGAGCAUGUAUUUCCUUCCU-UUCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCACUCUGAUUGAAAGUC --------(((((((...........--.....(((........).)).((((((........-((((((((....))))))))..........))))))...)))))))..... ( -23.47) >DroAna_CAF1 21487 99 + 1 ----------AGUCCU--C----CUCCUCGGCGGAAAACCAGUCGAGCUUGUAUUUCCUUCCUCUUCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCACUCUGAUUGAAAGUC ----------..(((.--(----(.....)).)))....((((((((..((((((.........((((((((....))))))))..........))))))))).)))))...... ( -28.31) >DroPer_CAF1 24883 100 + 1 --------UUAGUCCUUCA----GUA--CGUCAGUCCGCCAGUGGAGCAUGUAUUUCCUGCCU-UUCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCACUCUGAUUGAAAGUC --------.......((((----...--.(((((...((...((((((..(((.....)))..-((((((((....)))))))).....))))))..))...))))))))).... ( -26.90) >consensus __________AGUCCU__A____GUC__CGUCAGACAUCCAGUCGAGCAUGUAUUUCCUUCCUCUUCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCACUCUGAUUGAAAGUC .......................................((((((((..((((((.........((((((((....))))))))..........))))))))).)))))...... (-19.67 = -20.39 + 0.72)

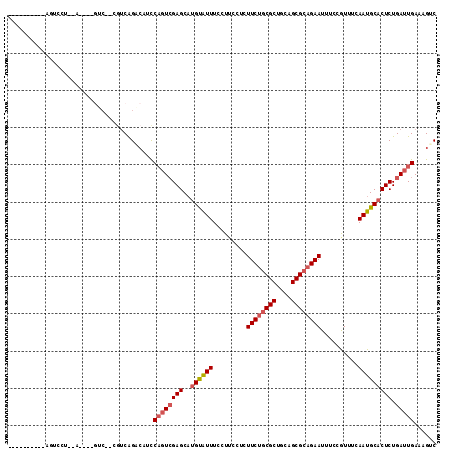
| Location | 10,684,839 – 10,684,937 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.79 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -21.20 |
| Energy contribution | -22.37 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.924942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10684839 98 + 20766785 --UCCAGUCGAGCAUGCAUUUCCUUCCUCUUCUGCGCUGCAGCGCAGAAUUUCCAUUCCAAUGCACUCUGAUUGAAAGUCUUACGUGCUG--------------------UGUGGGUCCU --..((((((((..((((((.........((((((((....))))))))..........))))))))).)))))...(.((((((.....--------------------)))))).).. ( -28.71) >DroVir_CAF1 36272 97 + 1 --UUCCCUCGAGCAUGUAUUUCCUUCCU--UCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCACCCUGAUUGAAAGUCUUACGUACUUA-------------------UGUGGCCCUU --..(((..(((.(((((.........(--(((((((....))))))))......(((((((........)))))))....))))).))).-------------------.).))..... ( -23.30) >DroGri_CAF1 27554 100 + 1 AGUUCACACGUGCAUGUAUUUCCUUCCU-UUCUGCGCUGCAGCGCAGAAUUUCCGUUUCGGUGCACUCUGAUUGAAAGUCUUACGUACUUA-------------------UGUGGCCCUU .(..((((.((((...............-((((((((....))))))))((((....((((......))))..)))).......))))...-------------------))))..)... ( -25.50) >DroWil_CAF1 31333 116 + 1 --GCCAGUCGAGCAUGUGUUUCCUUCCUUUUCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCCCUCUGAUUGAAAGACUUACGUGCUCAAGGACCUCCAACCCUGCAGUGUUGGCC-- --(((((.((((((((((..((.......((((((((....))))))))......(((((((........)))))))))..)))))))))..((....))...........)))))).-- ( -40.10) >DroAna_CAF1 21508 98 + 1 --ACCAGUCGAGCUUGUAUUUCCUUCCUCUUCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCACUCUGAUUGAAAGUCUUACGUGCUU--------------------UUUGGUUAUU --(((((..((((.((((...........((((((((....))))))))......(((((((........)))))))....)))).))))--------------------.))))).... ( -27.80) >DroPer_CAF1 24906 98 + 1 --GCCAGUGGAGCAUGUAUUUCCUGCCU-UUCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCACUCUGAUUGAAAGUCUUACGUGGUG------C-------------UCUGUGUCUG --(((((.(.(.((((((..........-((((((((....))))))))......(((((((........)))))))....)))))).).------)-------------.))).))... ( -29.20) >consensus __UCCAGUCGAGCAUGUAUUUCCUUCCU_UUCUGCGCUGCAGCGCAGAAUUUCCGUUUCAAUGCACUCUGAUUGAAAGUCUUACGUGCUU____________________UGUGGGCCUU .........(((((((((...........((((((((....))))))))......(((((((........)))))))....))))))))).............................. (-21.20 = -22.37 + 1.17)


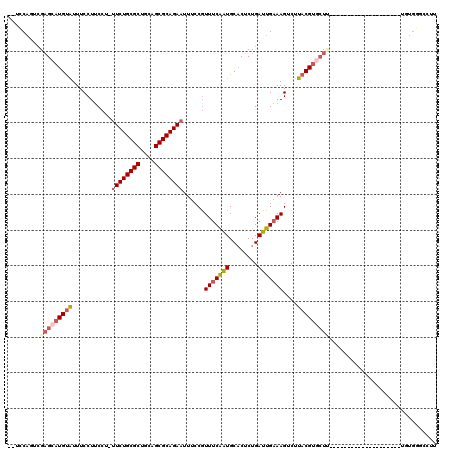
| Location | 10,684,839 – 10,684,937 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.79 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.18 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10684839 98 - 20766785 AGGACCCACA--------------------CAGCACGUAAGACUUUCAAUCAGAGUGCAUUGGAAUGGAAAUUCUGCGCUGCAGCGCAGAAGAGGAAGGAAAUGCAUGCUCGACUGGA-- .....(((..--------------------((((.(....).............(((((((..........((((((((....)))))))).........)))))))))).)..))).-- ( -26.91) >DroVir_CAF1 36272 97 - 1 AAGGGCCACA-------------------UAAGUACGUAAGACUUUCAAUCAGGGUGCAUUGAAACGGAAAUUCUGCGCUGCAGCGCAGA--AGGAAGGAAAUACAUGCUCGAGGGAA-- .....((...-------------------..((((.(((...(((((..((((......))))........((((((((....)))))))--))))))....))).))))....))..-- ( -25.50) >DroGri_CAF1 27554 100 - 1 AAGGGCCACA-------------------UAAGUACGUAAGACUUUCAAUCAGAGUGCACCGAAACGGAAAUUCUGCGCUGCAGCGCAGAA-AGGAAGGAAAUACAUGCACGUGUGAACU ..((..((((-------------------(((((.(....))))).........((((((((...)))...((((((((....))))))))-..............))))))))))..)) ( -31.00) >DroWil_CAF1 31333 116 - 1 --GGCCAACACUGCAGGGUUGGAGGUCCUUGAGCACGUAAGUCUUUCAAUCAGAGGGCAUUGAAACGGAAAUUCUGCGCUGCAGCGCAGAAAAGGAAGGAAACACAUGCUCGACUGGC-- --..(((((........))))).((((...(((((.((...((((((..((((......))))........((((((((....))))))))...))))))...)).)))))))))...-- ( -37.40) >DroAna_CAF1 21508 98 - 1 AAUAACCAAA--------------------AAGCACGUAAGACUUUCAAUCAGAGUGCAUUGAAACGGAAAUUCUGCGCUGCAGCGCAGAAGAGGAAGGAAAUACAAGCUCGACUGGU-- ....((((..--------------------.(((..(((...(((((..((...((........)).))..((((((((....))))))))..)))))....)))..)))....))))-- ( -25.40) >DroPer_CAF1 24906 98 - 1 CAGACACAGA-------------G------CACCACGUAAGACUUUCAAUCAGAGUGCAUUGAAACGGAAAUUCUGCGCUGCAGCGCAGAA-AGGCAGGAAAUACAUGCUCCACUGGC-- (((...(((.-------------(------(((..(....)....((.....)))))).)))....(((..((((((((....))))))))-..((((......).)))))).)))..-- ( -27.70) >consensus AAGACCCACA____________________AAGCACGUAAGACUUUCAAUCAGAGUGCAUUGAAACGGAAAUUCUGCGCUGCAGCGCAGAA_AGGAAGGAAAUACAUGCUCGACUGGA__ ................................(((.(((...(((((..((((......))))........((((((((....))))))))..)))))....))).)))........... (-19.82 = -20.18 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:16 2006